Transcriptome analysis of buds and leaves using 454 pyrosequencing to discover genes associated with the biosynthesis of active ingredients in Lonicera japonica Thunb
- PMID: 23638167
- PMCID: PMC3636143
- DOI: 10.1371/journal.pone.0062922
Transcriptome analysis of buds and leaves using 454 pyrosequencing to discover genes associated with the biosynthesis of active ingredients in Lonicera japonica Thunb
Abstract
Background: Lonicera japonica Thunb. is a plant used in traditional Chinese medicine known for its anti-inflammatory, anti-oxidative, anti-carcinogenic, and antiviral pharmacological properties. The major active secondary metabolites of this plant are chlorogenic acid (CGA) and luteoloside. While the biosynthetic pathways of these metabolites are relatively well known, the genetic information available for this species, especially the biosynthetic pathways of its active ingredients, is limited.
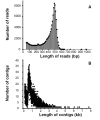
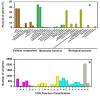
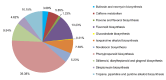
Methodology/principal findings: We obtained one million reads (average length of 400 bp) in a whole sequence run using a Roche/454 GS FLX titanium platform. Altogether, 85.69% of the unigenes covering the entire life cycle of the plant were annotated and 325 unigenes were assigned to secondary metabolic pathways. Moreover, 2039 unigenes were predicted as transcription factors. Nearly all of the possible enzymes involved in the biosynthesis of CGA and luteoloside were discovered in L. japonica. Three hydroxycinnamoyl transferase genes, including two hydroxycinnamoyl-CoA quinate hydroxycinnamoyl transferase genes and one hydroxycinnamoyl-CoA shikimate/quinate hydroxycinnamoyl transferase (HCT) gene featuring high similarity to known genes from other species, were cloned. The HCT gene was discovered for the first time in L. japonica. In addition, 188 candidate cytochrome P450 unigenes and 245 glycosyltransferase unigenes were found in the expressed sequence tag (EST) dataset.
Conclusion: This study provides a high quality EST database for L. japonica by 454 pyrosequencing. Based on the EST annotation, a set of putative genes involved in CGA and luteoloside biosynthetic pathways were discovered. The database serves as an important source of public information on genetic markers, gene expression, genomics, and functional genomics in L. japonica.
Conflict of interest statement
Figures


References
-
- Schierenbeck KA (2004) Japanese Honeysuckle (Lonicera japonica) as an invasive species; history, ecology, and context. CRIT REV PLANT SCI 23: 391–400.
-
- Chang WC, Hsu FL (1992) Inhibition of platelet activation and endothelial cell injury by polyphenolic compounds isolated from Lonicera japonica Thunb. PLEFA 45: 307–312. - PubMed
-
- Clifford MN (2000) Chlorogenic acids and other cinnamates-nature, occurrence, dietary burden, absorption and metabolism. J Sci Food Agr 80: 1033–1043.
-
- Lu HT, Jiang Y, Chen F (2004) Application of preparative high-speed counter-current chromatography for separation of chlorogenic acid from Flos Lonicerae. J Chromatogr A 1026: 185–190. - PubMed
-
- Inagaki I, Sakushima A, Hisada S, Nishibe S, Morita N (1974) Comparison of lonicerin and veronicastroside. Yakugaku Zasshi 94: 524–525. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

