A metagenomic insight into freshwater methane-utilizing communities and evidence for cooperation between the Methylococcaceae and the Methylophilaceae
- PMID: 23638358
- PMCID: PMC3628875
- DOI: 10.7717/peerj.23
A metagenomic insight into freshwater methane-utilizing communities and evidence for cooperation between the Methylococcaceae and the Methylophilaceae
Abstract
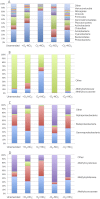
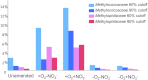
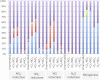
We investigated microbial communities active in methane oxidation in lake sediment at different oxygen tensions and their response to the addition of nitrate, via stable isotope probing combined with deep metagenomic sequencing. Communities from a total of four manipulated microcosms were analyzed, supplied with (13)C-methane in, respectively, ambient air, ambient air with the addition of nitrate, nitrogen atmosphere and nitrogen atmosphere with the addition of nitrate, and these were compared to the community from an unamended sediment sample. We found that the major group involved in methane oxidation in both aerobic and microaerobic conditions were members of the family Methylococcaceae, dominated by species of the genus Methylobacter, and these were stimulated by nitrate in aerobic but not microaerobic conditions. In aerobic conditions, we also noted a pronounced response to both methane and nitrate by members of the family Methylophilaceae that are non-methane-oxidizing methylotrophs, and predominantly by the members of the genus Methylotenera. The relevant abundances of the Methylococcaceae and the Methylophilaceae and their coordinated response to methane and nitrate suggest that these species may be engaged in cooperative behavior, the nature of which remains unknown.
Keywords: Lake sediment; Metagenomics; Methane; Methylococcaceae; Methylophilaceae; Methylotrophy; Microbial community; Nitrate.
Figures

References
-
- Anthony C. The biochemistry of methylotrophs. London: Academic Press; 1982.
-
- Beck DAC, Hendrickson EL, Vorobev A, Wang T, Lim S, Kalyuzhnaya MG, Lidstrom ME, Hackett M, Chistoserdova L. An integrated proteomics/transcriptomics approach points to oxygen as the main electron sink for methanol metabolism in Methylotenera mobilis. Journal of Bacteriology. 2011;193:4758–4765. doi: 10.1128/JB.05375-11. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

