LSSP-PCR of Trypanosoma cruzi: how the single primer sequence affects the kDNA signature
- PMID: 23639061
- PMCID: PMC3653686
- DOI: 10.1186/1756-0500-6-174
LSSP-PCR of Trypanosoma cruzi: how the single primer sequence affects the kDNA signature
Abstract
Background: Low-stringency single specific primer PCR (LSSP-PCR) is a highly sensitive and discriminating technique that has been extensively used to genetically characterize Trypanosoma cruzi populations in the presence of large amounts of host DNA. To ensure high sensitivity, in most T. cruzi studies, the variable regions of the naturally amplified kinetoplast DNA (kDNA) minicircles were targeted, and this method translated the intraspecific polymorphisms of these molecules into specific and reproducible kDNA signatures. Although the LSSP-PCR technique is reproducible under strict assay conditions, the complex banding pattern generated can be significantly altered by even a single-base change in the target DNA. Our survey of the literature identified eight different primers with similar, if not identical, names that have been used for kDNA amplification and LSSP-PCR of T. cruzi. Although different primer sequences were used in these studies, many of the authors cited the same reference report to justify their primer choice. We wondered whether these changes in the primer sequence could affect also the parasite LSSP-PCR profiles.
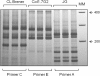
Findings: To answer this question we compared the kDNA signatures obtained from three different and extensively studied T. cruzi populations with the eight primers found in the literature. Our results clearly demonstrate that even minimal modifications in the oligonucleotide sequences, especially in the 3' or 5' end, can significantly change the kDNA signature of a T. cruzi strain.
Conclusions: These results highlight the necessity of careful preservation of primer nomenclature and sequence when reproducing an LSSP-PCR work to avoid confusion and allow comparison of results among different laboratories.
Figures


Similar articles
-
High variability of Colombian Trypanosoma cruzi lineage I stocks as revealed by low-stringency single primer-PCR minicircle signatures.Acta Trop. 2006 Nov;100(1-2):110-8. doi: 10.1016/j.actatropica.2006.10.003. Epub 2006 Nov 13. Acta Trop. 2006. PMID: 17101108
-
Kinetoplast DNA signatures of Trypanosoma cruzi strains obtained directly from infected tissues.Am J Pathol. 1996 Dec;149(6):2153-9. Am J Pathol. 1996. PMID: 8952547 Free PMC article.
-
Geographical clustering of Trypanosoma cruzi I groups from Colombia revealed by low-stringency single specific primer-PCR of the intergenic regions of spliced-leader genes.Parasitol Res. 2009 Jan;104(2):399-410. doi: 10.1007/s00436-008-1212-0. Epub 2008 Oct 11. Parasitol Res. 2009. PMID: 18850114
-
[Genetic variability of Trypanosoma cruzi in blood and organs of infected mice determined by LSSP-PCR].Biomedica. 2005 Mar;25(1):76-86. Biomedica. 2005. PMID: 15962904 Spanish.
-
Mitochondrial and satellite real time-PCR for detecting T. cruzi DTU II strain in blood and organs of experimentally infected mice presenting different levels of parasite load.Exp Parasitol. 2019 May;200:13-15. doi: 10.1016/j.exppara.2019.03.007. Epub 2019 Mar 20. Exp Parasitol. 2019. PMID: 30904696 Review.
Cited by
-
Intra-Discrete Typing Unit TcV Genetic Variability of Trypanosoma cruzi in Chronic Chagas' Disease Bolivian Immigrant Patients in Barcelona, Spain.Front Cardiovasc Med. 2021 May 20;8:665624. doi: 10.3389/fcvm.2021.665624. eCollection 2021. Front Cardiovasc Med. 2021. PMID: 34095255 Free PMC article.
-
Between a bug and a hard place: Trypanosoma cruzi genetic diversity and the clinical outcomes of Chagas disease.Expert Rev Anti Infect Ther. 2015 Aug;13(8):995-1029. doi: 10.1586/14787210.2015.1056158. Expert Rev Anti Infect Ther. 2015. PMID: 26162928 Free PMC article. Review.
References
-
- Morel C, Simpson L. Characterization of pathogenic trypanosomatidae by restriction endonuclease fingerprinting of kinetoplast DNA minicircles. Am J Trop Med Hyg. 1980;29:1070–1074. - PubMed
-
- Dias Neto E, Steindel M, Passos LK, de Souza CP, Rollinson D, Katz N, Romanha AJ, Pena SD, Simpson AJ. The use of RAPDs for the study of the genetic diversity of Schistosoma mansoni and Trypanosoma cruzi. EXS. 1993;67:339–345. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

