Within-host and between-host evolutionary rates across the HIV-1 genome
- PMID: 23639104
- PMCID: PMC3685529
- DOI: 10.1186/1742-4690-10-49
Within-host and between-host evolutionary rates across the HIV-1 genome
Abstract
Background: HIV evolves rapidly at the epidemiological level but also at the within-host level. The virus' within-host evolutionary rates have been argued to be much higher than its between-host evolutionary rates. However, this conclusion relies on analyses of a short portion of the virus envelope gene. Here, we study in detail these evolutionary rates across the HIV genome.
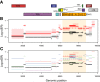
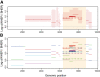
Results: We build phylogenies using a relaxed molecular clock assumption to estimate evolutionary rates in different regions of the HIV genome. We find that these rates vary strongly across the genome, with higher rates in the envelope gene (env). Within-host evolutionary rates are consistently higher than between-host rates throughout the HIV genome. This difference is significantly more pronounced in env. Finally, we find weak differences between overlapping and non-overlapping regions.
Conclusions: We provide a genome-wide overview of the differences in the HIV rates of molecular evolution at the within- and between-host levels. Contrary to hepatitis C virus, where differences are only located in the envelope gene, within-host evolutionary rates are higher than between-host evolutionary rates across the whole HIV genome. This supports the hypothesis that HIV strains that are less adapted to the host have an advantage during transmission. The most likely mechanism for this is storage and then preferential transmission of viruses in latent T-cells. These results shed a new light on the role of the transmission bottleneck in the evolutionary dynamics of HIV.
Figures



References
-
- Shankarappa R, Margolick JB, Gange SJ, Rodrigo AG, Upchurch D, Farzadegan H, Gupta P, Rinaldo CR, Learn GH, He X, Huang XL, Mullins JI. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J Virol. 1999;73(12):10489–10502. - PMC - PubMed
-
- Lemey P, Rambaut A, Pybus OG. HIV evolutionary dynamics within and among hosts. AIDS Rev. 2006;8(3):125–140. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

