4FISH-IF, a four-color dual-gene FISH combined with p63 immunofluorescence to evaluate NKX3.1 and MYC status in prostate cancer
- PMID: 23640976
- PMCID: PMC3707357
- DOI: 10.1369/0022155413490946
4FISH-IF, a four-color dual-gene FISH combined with p63 immunofluorescence to evaluate NKX3.1 and MYC status in prostate cancer
Abstract
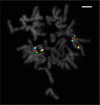
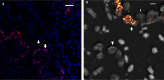
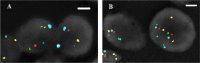
NKX3.1 allelic loss and MYC amplification are common events during prostate cancer progression and have been recognized as potential prognostic factors in prostate cancer after radical prostatectomy or precision radiotherapy. We have developed a 4FISH-IF assay (a dual-gene fluorescence in situ hybridization combined with immunofluorescence) to measure both NKX3.1 and MYC status on the same slide. The 4FISH-IF assay contains four probes complementary to chromosome 8 centromere, 8p telomere, 8p21, and 8q24, as well as an antibody targeting the basal cell marker p63 visualized by immunofluorescence. The major advantages of the 4FISH-IF include the distinction between benign and malignant glands directly on the 4FISH-IF slide and the control of truncation artifact. Importantly, this specialized and innovative combined multiprobe and immunofluorescence technique can be performed on diagnostic biopsy specimens, increasing its clinical relevance. Moreover, the assay can be easily performed in a standard clinical molecular pathology laboratory. Globally, the use of 4FISH-IF decreases analytic time, increases confidence in obtained results, and maintains the tissue morphology of the diagnostic specimen.
Keywords: MYC; NKX3.1; biopsy; fluorescence in situ hybridization; immunofluorescence; prostate cancer.
Conflict of interest statement
Figures
References
-
- Asatiani E, Huang WX, Wang A, Rodriguez Ortner E, Cavalli LR, Haddad BR, Gelmann EP. 2005. Deletion, methylation, and expression of the NKX3.1 suppressor gene in primary human prostate cancer. Cancer Res. 65:1164–1173 - PubMed
-
- Bethel CR, Faith D, Li X, Guan B, Hicks JL, Lan F, Jenkins RB, Bieberich CJ, De Marzo AM. 2006. Decreased NKX3.1 protein expression in focal prostatic atrophy, prostatic intraepithelial neoplasia, and adenocarcinoma: association with Gleason score and chromosome 8p deletion. Cancer Res. 66:10683–10690 - PubMed
-
- Bowen C, Bubendorf L, Voeller HJ, Slack R, Willi N, Sauter G, Gasser TC, Koivisto P, Lack EE, Kononen J, et al. 2000. Loss of NKX3.1 expression in human prostate cancers correlates with tumor progression. Cancer Res. 60:6111–6115 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

