Mutations in epigenetic modifiers in the pathogenesis and therapy of acute myeloid leukemia
- PMID: 23640996
- PMCID: PMC3643757
- DOI: 10.1182/blood-2013-01-451781
Mutations in epigenetic modifiers in the pathogenesis and therapy of acute myeloid leukemia
Abstract
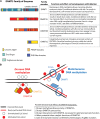
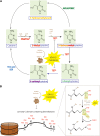
Recent studies of the spectrum of somatic genetic alterations in acute myeloid leukemia (AML) have identified frequent somatic mutations in genes that encode proteins important in the epigenetic regulation of gene transcription. This includes proteins involved in the modification of DNA cytosine residues and enzymes which catalyze posttranslational modifications of histones. Here we describe the clinical, biological, and therapeutic relevance of mutations in epigenetic regulators in AML. In particular, we focus on the role of loss-of-function mutations in TET2, gain-of-function mutations in IDH1 and IDH2, and loss-of-function mutations in ASXL1 and mutations of unclear impact in DNMT3A in AML pathogenesis and therapy. Multiple studies have consistently identified that mutations in these genes have prognostic relevance, particularly in intermediate-risk AML patients, arguing for inclusion of mutational testing of these genetic abnormalities in routine clinical practice. Moreover, biochemical, biological, and epigenomic analyses of the effects of these mutations have informed the development of novel therapies which target pathways deregulated by these mutations. Our understanding of the effects of these mutations on hematopoiesis and potential for therapeutic targeting of specific AML subsets is also reviewed here.
Figures
References
-
- Chou WC, Chou SC, Liu CY, et al. TET2 mutation is an unfavorable prognostic factor in acute myeloid leukemia patients with intermediate-risk cytogenetics. Blood. 2011;118(14):3803–3810. - PubMed
-
- Damm F, Thol F, Hollink I, et al. Prevalence and prognostic value of IDH1 and IDH2 mutations in childhood AML: a study of the AML-BFM and DCOG study groups. Leukemia. 2011;25(11):1704–1710. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

