Synapsable quadruplex-mediated fibers
- PMID: 23641903
- PMCID: PMC3655031
- DOI: 10.1186/1556-276X-8-210
Synapsable quadruplex-mediated fibers
Abstract
We have fabricated a DNA-based nanofiber created by self-assembly of guanine quadruplex (Hoogsteen base pairing) and double-stranded DNA (Watson-Crick base pairing). When duplexes containing a long stretch of contiguous guanines and single-stranded overhangs are incubated in potassium-containing buffer, the preformed duplexes create high molecular weight species that contain quadruplexes. In addition to observation of these larger species by gel electrophoresis, solutions were analyzed by atomic force microscopy to reveal nanofibers. Analysis of the atomic force microscopy images indicates that fibers form with lengths ranging from 250 to 2,000 nm and heights from 0.45 to 4.0 nm. This work is a first step toward the creation of new structurally heterogeneous (quadruplex/duplex), yet controllable, DNA-based materials exhibiting novel properties suitable for a diverse array of nanotechnology applications.
Figures


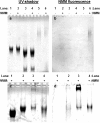
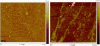
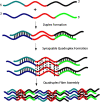
Similar articles
-
Charge conduction properties of a parallel-stranded DNA G-quadruplex: implications for chromosomal oxidative damage.Biochemistry. 2009 Jul 28;48(29):6794-804. doi: 10.1021/bi9007484. Biochemistry. 2009. PMID: 19530739
-
Formation and Nanomechanical Properties of Silver-Mediated Guanine DNA Duplexes in Aqueous Solution.ACS Nano. 2024 Jan 30;18(4):3002-3010. doi: 10.1021/acsnano.3c08008. Epub 2024 Jan 16. ACS Nano. 2024. PMID: 38227309 Free PMC article.
-
Single strand targeted triplex-formation. Destabilization of guanine quadruplex structures by foldback triplex-forming oligonucleotides.Nucleic Acids Res. 1995 Mar 25;23(6):1068-74. doi: 10.1093/nar/23.6.1068. Nucleic Acids Res. 1995. PMID: 7537368 Free PMC article.
-
Watson-Crick versus Hoogsteen Base Pairs: Chemical Strategy to Encode and Express Genetic Information in Life.Acc Chem Res. 2021 May 4;54(9):2110-2120. doi: 10.1021/acs.accounts.0c00734. Epub 2021 Feb 16. Acc Chem Res. 2021. PMID: 33591181 Review.
-
Thermodynamic and kinetic characterization of the dissociation and assembly of quadruplex nucleic acids.Biopolymers. 2000-2001;56(3):147-94. doi: 10.1002/1097-0282(2000/2001)56:3<147::AID-BIP10011>3.0.CO;2-N. Biopolymers. 2000. PMID: 11745110 Review.
Cited by
-
Spontaneous DNA Synapsis by Forming Noncanonical Intermolecular Structures.Polymers (Basel). 2022 May 23;14(10):2118. doi: 10.3390/polym14102118. Polymers (Basel). 2022. PMID: 35632001 Free PMC article.
-
Development of moisture-proof polydimethylsiloxane/aluminum oxide film and stability improvement of perovskite solar cells using the film.RSC Adv. 2019 Apr 15;9(21):11737-11744. doi: 10.1039/c9ra01107b. eCollection 2019 Apr 12. RSC Adv. 2019. PMID: 35517001 Free PMC article.
-
Visualization of G-Quadruplexes, i-Motifs and Their Associates.Acta Naturae. 2022 Jul-Sep;14(3):4-18. doi: 10.32607/actanaturae.11705. Acta Naturae. 2022. PMID: 36348720 Free PMC article.
References
-
- Sugimoto N. Designable DNA functions toward new nanobiotechnology. Bull Chem Soc Jpn. 2009;8:1–10. doi: 10.1246/bcsj.82.1. - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources

