Molecular diversity and population structure at the Cytochrome P450 3A5 gene in Africa
- PMID: 23641907
- PMCID: PMC3655848
- DOI: 10.1186/1471-2156-14-34
Molecular diversity and population structure at the Cytochrome P450 3A5 gene in Africa
Abstract
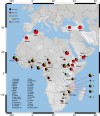
Background: Cytochrome P450 3A5 (CYP3A5) is an enzyme involved in the metabolism of many therapeutic drugs. CYP3A5 expression levels vary between individuals and populations, and this contributes to adverse clinical outcomes. Variable expression is largely attributed to four alleles, CYP3A5*1 (expresser allele); CYP3A5*3 (rs776746), CYP3A5*6 (rs10264272) and CYP3A5*7 (rs41303343) (low/non-expresser alleles). Little is known about CYP3A5 variability in Africa, a region with considerable genetic diversity. Here we used a multi-disciplinary approach to characterize CYP3A5 variation in geographically and ethnically diverse populations from in and around Africa, and infer the evolutionary processes that have shaped patterns of diversity in this gene. We genotyped 2538 individuals from 36 diverse populations in and around Africa for common low/non-expresser CYP3A5 alleles, and re-sequenced the CYP3A5 gene in five Ethiopian ethnic groups. We estimated the ages of low/non-expresser CYP3A5 alleles using a linked microsatellite and assuming a step-wise mutation model of evolution. Finally, we examined a hypothesis that CYP3A5 is important in salt retention adaptation by performing correlations with ecological data relating to aridity for the present day, 10,000 and 50,000 years ago.
Results: We estimate that ~43% of individuals within our African dataset express CYP3A5, which is lower than previous independent estimates for the region. We found significant intra-African variability in CYP3A5 expression phenotypes. Within Africa the highest frequencies of high-activity alleles were observed in equatorial and Niger-Congo speaking populations. Ethiopian allele frequencies were intermediate between those of other sub-Saharan African and non-African groups. Re-sequencing of CYP3A5 identified few additional variants likely to affect CYP3A5 expression. We estimate the ages of CYP3A5*3 as ~76,400 years and CYP3A5*6 as ~218,400 years. Finally we report that global CYP3A5 expression levels correlated significantly with aridity measures for 10,000 [Spearmann's Rho= -0.465, p=0.004] and 50,000 years ago [Spearmann's Rho= -0.379, p=0.02].
Conclusions: Significant intra-African diversity at the CYP3A5 gene is likely to contribute to multiple pharmacogenetic profiles across the continent. Significant correlations between CYP3A5 expression phenotypes and aridity data are consistent with a hypothesis that the enzyme is important in salt-retention adaptation.
Figures

References
-
- Coleman R. Disease burden in sub-Saharan Africa. Lancet. 1998;351(9110):1208. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

