DNA branch migration reactions through photocontrollable toehold formation
- PMID: 23642046
- PMCID: PMC3742003
- DOI: 10.1021/ja4018495
DNA branch migration reactions through photocontrollable toehold formation
Abstract
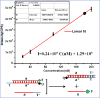
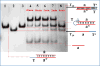
Strand displacement cascades are commonly used to make dynamically assembled structures. Particularly, the concept of "toehold-mediated DNA branch migration reactions" has attracted considerable attention in relation to dynamic DNA nanostructures. However, it is a challenge to obtain and control the formation of pure 1:1 ratio DNA duplexes with toehold structures. Here, for the first time, we report a photocontrolled toehold formation method, which is based on the photocleavage of 2-nitrobenzyl linker-embedded DNA hairpin precursor structures. UV light irradiation (λ ≈ 365 nm) of solutions containing these DNA hairpin structures causes the complete cleavage of the nitrobenzyl linker, and pure 1:1 DNA duplexes with toehold structures are easily formed. Our experimental results indicate that the amount of toehold can be controlled by simply changing the dose of UV irradiation and that the resulting toehold structures can be used for subsequent toehold-mediated DNA branch migration reactions, e.g., DNA hybridization chain reactions. This newly established method will find broad application in the construction of light-powered, controllable, and dynamic DNA nanostructures or large-scale DNA circuits.
Figures




Similar articles
-
Toehold clipping: A mechanism for remote control of DNA strand displacement.Nucleic Acids Res. 2023 May 8;51(8):4055-4063. doi: 10.1093/nar/gkac1152. Nucleic Acids Res. 2023. PMID: 36477864 Free PMC article.
-
Regulation of DNA Strand Displacement Using an Allosteric DNA Toehold.J Am Chem Soc. 2016 Oct 26;138(42):14076-14082. doi: 10.1021/jacs.6b08794. Epub 2016 Oct 13. J Am Chem Soc. 2016. PMID: 27704809
-
Multicolor and erasable DNA photolithography.ACS Nano. 2014 Jul 22;8(7):6849-55. doi: 10.1021/nn5024472. Epub 2014 Jul 7. ACS Nano. 2014. PMID: 24988147 Free PMC article.
-
Principles and Applications of Nucleic Acid Strand Displacement Reactions.Chem Rev. 2019 May 22;119(10):6326-6369. doi: 10.1021/acs.chemrev.8b00580. Epub 2019 Feb 4. Chem Rev. 2019. PMID: 30714375 Review.
-
DNA Strand Displacement Reaction: A Powerful Tool for Discriminating Single Nucleotide Variants.Top Curr Chem (Cham). 2020 Jan 2;378(1):10. doi: 10.1007/s41061-019-0274-z. Top Curr Chem (Cham). 2020. PMID: 31894426 Review.
Cited by
-
Nucleic acid based logical systems.Chemistry. 2014 May 12;20(20):5866-73. doi: 10.1002/chem.201304891. Epub 2014 Apr 1. Chemistry. 2014. PMID: 24692306 Free PMC article.
-
Controllable DNA strand displacement by independent metal-ligand complexation.Chem Sci. 2021 May 18;12(25):8698-8705. doi: 10.1039/d1sc01041g. eCollection 2021 Jul 1. Chem Sci. 2021. PMID: 34257868 Free PMC article.
-
Expanding Catch and Release DNA Decoy (CRDD) Technology with Pyrimidine Mimics.Chemistry. 2022 Oct 18;28(58):e202201355. doi: 10.1002/chem.202201355. Epub 2022 Aug 25. Chemistry. 2022. PMID: 35849314 Free PMC article.
-
DNA-Based Dynamic Reaction Networks.Trends Biochem Sci. 2018 Jul;43(7):547-560. doi: 10.1016/j.tibs.2018.04.010. Epub 2018 May 21. Trends Biochem Sci. 2018. PMID: 29793809 Free PMC article. Review.
-
Signal amplified colorimetric nucleic acid detection based on autocatalytic hairpin assembly.RSC Adv. 2024 May 28;14(24):17152-17157. doi: 10.1039/d4ra01982b. eCollection 2024 May 22. RSC Adv. 2024. PMID: 38808241 Free PMC article.
References
-
- Lee CS, Davis RW, Davidson N. J Mol Biol. 1970;48:1–22. - PubMed
-
- Seeman NC. Angew Chem Int Edit. 1998;37:3220–3238. - PubMed
-
- Seeman N. C Nature. 2003;421:427–431. - PubMed
-
- Yurke B, Turberfield AJ, Mills AP, Simmel FC, Neumann JL. Nature. 2000;406:605–608. - PubMed
-
- Bath J, Turberfield AJ. Nat Nanotechnol. 2007;2:275–284. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

