Lamin A/C and emerin regulate MKL1-SRF activity by modulating actin dynamics
- PMID: 23644458
- PMCID: PMC3666313
- DOI: 10.1038/nature12105
Lamin A/C and emerin regulate MKL1-SRF activity by modulating actin dynamics
Abstract
Laminopathies, caused by mutations in the LMNA gene encoding the nuclear envelope proteins lamins A and C, represent a diverse group of diseases that include Emery-Dreifuss muscular dystrophy (EDMD), dilated cardiomyopathy (DCM), limb-girdle muscular dystrophy, and Hutchison-Gilford progeria syndrome. Most LMNA mutations affect skeletal and cardiac muscle by mechanisms that remain incompletely understood. Loss of structural function and altered interaction of mutant lamins with (tissue-specific) transcription factors have been proposed to explain the tissue-specific phenotypes. Here we report in mice that lamin-A/C-deficient (Lmna(-/-)) and Lmna(N195K/N195K) mutant cells have impaired nuclear translocation and downstream signalling of the mechanosensitive transcription factor megakaryoblastic leukaemia 1 (MKL1), a myocardin family member that is pivotal in cardiac development and function. Altered nucleo-cytoplasmic shuttling of MKL1 was caused by altered actin dynamics in Lmna(-/-) and Lmna(N195K/N195K) mutant cells. Ectopic expression of the nuclear envelope protein emerin, which is mislocalized in Lmna mutant cells and also linked to EDMD and DCM, restored MKL1 nuclear translocation and rescued actin dynamics in mutant cells. These findings present a novel mechanism that could provide insight into the disease aetiology for the cardiac phenotype in many laminopathies, whereby lamin A/C and emerin regulate gene expression through modulation of nuclear and cytoskeletal actin polymerization.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
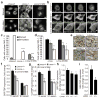

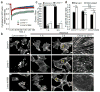

Comment in
-
SRF regulation - actin branches out.Nat Rev Mol Cell Biol. 2014 Jun;15(6):368. doi: 10.1038/nrm3803. Epub 2014 Apr 30. Nat Rev Mol Cell Biol. 2014. PMID: 24781641 No abstract available.
References
-
- Miralles F, Posern G, Zaromytidou AI, Treisman R. Actin dynamics control SRF activity by regulation of its coactivator MAL. Cell. 2003;113:329–342. S0092867403002782 [pii] - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

