Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy
- PMID: 23644491
- PMCID: PMC3667980
- DOI: 10.1038/ng.2628
Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy
Abstract
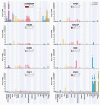
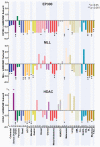
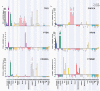
Subunits of mammalian SWI/SNF (mSWI/SNF or BAF) complexes have recently been implicated as tumor suppressors in human malignancies. To understand the full extent of their involvement, we conducted a proteomic analysis of endogenous mSWI/SNF complexes, which identified several new dedicated, stable subunits not found in yeast SWI/SNF complexes, including BCL7A, BCL7B and BCL7C, BCL11A and BCL11B, BRD9 and SS18. Incorporating these new members, we determined mSWI/SNF subunit mutation frequency in exome and whole-genome sequencing studies of primary human tumors. Notably, mSWI/SNF subunits are mutated in 19.6% of all human tumors reported in 44 studies. Our analysis suggests that specific subunits protect against cancer in specific tissues. In addition, mutations affecting more than one subunit, defined here as compound heterozygosity, are prevalent in certain cancers. Our studies demonstrate that mSWI/SNF is the most frequently mutated chromatin-regulatory complex (CRC) in human cancer, exhibiting a broad mutation pattern, similar to that of TP53. Thus, proper functioning of polymorphic BAF complexes may constitute a major mechanism of tumor suppression.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

