The transcriptional landscape of αβ T cell differentiation
- PMID: 23644507
- PMCID: PMC3660436
- DOI: 10.1038/ni.2590
The transcriptional landscape of αβ T cell differentiation
Abstract
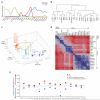
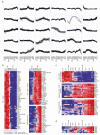
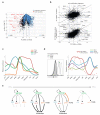
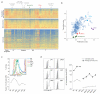
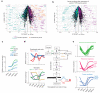
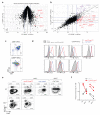
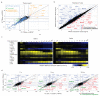
The differentiation of αβT cells from thymic precursors is a complex process essential for adaptive immunity. Here we exploited the breadth of expression data sets from the Immunological Genome Project to analyze how the differentiation of thymic precursors gives rise to mature T cell transcriptomes. We found that early T cell commitment was driven by unexpectedly gradual changes. In contrast, transit through the CD4(+)CD8(+) stage involved a global shutdown of housekeeping genes that is rare among cells of the immune system and correlated tightly with expression of the transcription factor c-Myc. Selection driven by major histocompatibility complex (MHC) molecules promoted a large-scale transcriptional reactivation. We identified distinct signatures that marked cells destined for positive selection versus apoptotic deletion. Differences in the expression of unexpectedly few genes accompanied commitment to the CD4(+) or CD8(+) lineage, a similarity that carried through to peripheral T cells and their activation, demonstrated by mass cytometry phosphoproteomics. The transcripts newly identified as encoding candidate mediators of key transitions help define the 'known unknowns' of thymocyte differentiation.
Figures

References
-
- Miller JF. The golden anniversary of the thymus. Nat Rev. Immunol. 2011;11:489–495. - PubMed
-
- von Boehmer H, Teh HS, Kisielow P. The thymus selects the useful, neglects the useless and destroys the harmful. Immunol. Today. 1989;10:57–61. - PubMed
-
- Jameson SC, Hogquist KA, Bevan MJ. Positive selection of thymocytes. Annu. Rev. Immunol. 1995;13:93–126. - PubMed
-
- Robey E. Regulation of T cell fate by Notch. Annu. Rev. Immunol. 1999;17:283–295. - PubMed
-
- Hogquist KA, Baldwin TA, Jameson SC. Central tolerance: learning self-control in the thymus. Nat. Rev. Immunol. 2005;5:772–782. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

