A genome-wide methylation study on obesity: differential variability and differential methylation
- PMID: 23644594
- PMCID: PMC3741222
- DOI: 10.4161/epi.24506
A genome-wide methylation study on obesity: differential variability and differential methylation
Abstract
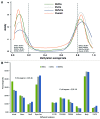
Besides differential methylation, DNA methylation variation has recently been proposed and demonstrated to be a potential contributing factor to cancer risk. Here we aim to examine whether differential variability in methylation is also an important feature of obesity, a typical non-malignant common complex disease. We analyzed genome-wide methylation profiles of over 470,000 CpGs in peripheral blood samples from 48 obese and 48 lean African-American youth aged 14-20 y old. A substantial number of differentially variable CpG sites (DVCs), using statistics based on variances, as well as a substantial number of differentially methylated CpG sites (DMCs), using statistics based on means, were identified. Similar to the findings in cancers, DVCs generally exhibited an outlier structure and were more variable in cases than in controls. By randomly splitting the current sample into a discovery and validation set, we observed that both the DVCs and DMCs identified from the first set could independently predict obesity status in the second set. Furthermore, both the genes harboring DMCs and the genes harboring DVCs showed significant enrichment of genes identified by genome-wide association studies on obesity and related diseases, such as hypertension, dyslipidemia, type 2 diabetes and certain types of cancers, supporting their roles in the etiology and pathogenesis of obesity. We generalized the recent finding on methylation variability in cancer research to obesity and demonstrated that differential variability is also an important feature of obesity-related methylation changes. Future studies on the epigenetics of obesity will benefit from both statistics based on means and statistics based on variances.
Keywords: African-Americans; epigenome-wide association study (EWAS); genome-wide association study (GWAS); methylation variation; obesity.
Figures




References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
