The octamer is the major form of CENP-A nucleosomes at human centromeres
- PMID: 23644596
- PMCID: PMC3760417
- DOI: 10.1038/nsmb.2562
The octamer is the major form of CENP-A nucleosomes at human centromeres
Abstract
The centromere is the chromosomal locus that ensures fidelity in genome transmission at cell division. Centromere protein A (CENP-A) is a histone H3 variant that specifies centromere location independently of DNA sequence. Conflicting evidence has emerged regarding the histone composition and stoichiometry of CENP-A nucleosomes. Here we show that the predominant form of the CENP-A particle at human centromeres is an octameric nucleosome. CENP-A nucleosomes are very highly phased on α-satellite 171-base-pair monomers at normal centromeres and also display strong positioning at neocentromeres. At either type of functional centromere, CENP-A nucleosomes exhibit similar DNA-wrapping behavior, as do octameric CENP-A nucleosomes reconstituted with recombinant components, having looser DNA termini than those on conventional nucleosomes containing canonical histone H3. Thus, the fundamental unit of the chromatin that epigenetically specifies centromere location in mammals is an octameric nucleosome with loose termini.
Figures

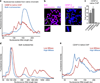
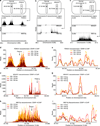

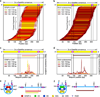
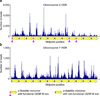

Comment in
-
Solo or doppio: how many CENP-As make a centromeric nucleosome?Nat Struct Mol Biol. 2013 Jun;20(6):648-50. doi: 10.1038/nsmb.2602. Nat Struct Mol Biol. 2013. PMID: 23739165 No abstract available.
References
-
- Cleveland DW, Mao Y, Sullivan KF. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell. 2003;112:407–421. - PubMed
-
- Mendiburo MJ, Padeken J, Fülöp S, Schepers A, Heun P. Drosophila CENH3 is sufficient for centromere formation. Science. 2011;334:686–690. - PubMed
-
- Warburton PE. Chromosomal dynamics of human neocentromere formation. Chromosome Res. 2004;12:617–626. - PubMed
Methods References
-
- Carruthers LM, Tse C, Walker KP, Hansen JC. Assembly of defined nucleosomal and chromatin arrays from pure components. Meth. Enzymol. 1999;304:19–35. - PubMed
-
- Luger K, Rechsteiner TJ, Richmond TJ. Preparation of nucleosome core particle from recombinant histones. Meth. Enzymol. 1999;304:3–19. - PubMed
-
- Lowary PT, Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 1998;276:19–42. - PubMed
-
- Huynh VAT, Robinson PJJ, Rhodes D. A Method for the In Vitro Reconstitution of a Defined ‘30nm’ Chromatin Fibre Containing Stoichiometric Amounts of the Linker Histone. J. Mol. Biol. 2005;345:957–968. - PubMed
-
- Brünger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

