First evidence of DNA methylation in insect Tribolium castaneum: environmental regulation of DNA methylation within heterochromatin
- PMID: 23644818
- PMCID: PMC3741223
- DOI: 10.4161/epi.24507
First evidence of DNA methylation in insect Tribolium castaneum: environmental regulation of DNA methylation within heterochromatin
Abstract
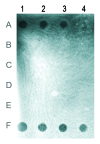
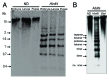
DNA methylation has been studied in many eukaryotic organisms, in particular vertebrates, and was implicated in developmental and phenotypic variations. Little is known about the role of DNA methylation in invertebrates, although insects are considered as excellent models for studying the evolution of DNA methylation. In the red flour beetle, Tribolium castaneum (Tenebrionidae, Coleoptera), no evidence of DNA methylation has been found till now. In this paper, a cytosine methylation in Tribolium castaneum embryos was detected by methylation sensitive restriction endonucleases and immuno-dot blot assay. DNA methylation in embryos is followed by a global demethylation in larvae, pupae and adults. DNA demethylation seems to proceed actively through 5-hydroxymethylcytosine, most probably by the action of TET enzyme. Bisulfite sequencing of a highly abundant satellite DNA located in pericentromeric heterochromatin revealed similar profile of cytosine methylation in adults and embryos. Cytosine methylation was not only restricted to CpG sites but was found at CpA, CpT and CpC sites. In addition, complete cytosine demethylation of heterochromatic satellite DNA was induced by heat stress. The results reveal existence of DNA methylation cycling in T. castaneum ranging from strong overall cytosine methylation in embryos to a weak DNA methylation in other developmental stages. Nevertheless, DNA methylation is preserved within heterochromatin during development, indicating its role in heterochromatin formation and maintenance. It is, however, strongly affected by heat stress, suggesting a role for DNA methylation in heterochromatin structure modulation during heat stress response.
Keywords: DNA methylation; Tribolium castaneum; heat shock response; heterochromatin; satellite DNA.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
