Insights into diphthamide, key diphtheria toxin effector
- PMID: 23645155
- PMCID: PMC3709272
- DOI: 10.3390/toxins5050958
Insights into diphthamide, key diphtheria toxin effector
Abstract
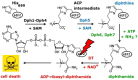
Diphtheria toxin (DT) inhibits eukaryotic translation elongation factor 2 (eEF2) by ADP-ribosylation in a fashion that requires diphthamide, a modified histidine residue on eEF2. In budding yeast, diphthamide formation involves seven genes, DPH1-DPH7. In an effort to further study diphthamide synthesis and interrelation among the Dph proteins, we found, by expression in E. coli and co-immune precipitation in yeast, that Dph1 and Dph2 interact and that they form a complex with Dph3. Protein-protein interaction mapping shows that Dph1-Dph3 complex formation can be dissected by progressive DPH1 gene truncations. This identifies N- and C-terminal domains on Dph1 that are crucial for diphthamide synthesis, DT action and cytotoxicity of sordarin, another microbial eEF2 inhibitor. Intriguingly, dph1 truncation mutants are sensitive to overexpression of DPH5, the gene necessary to synthesize diphthine from the first diphthamide pathway intermediate produced by Dph1-Dph3. This is in stark contrast to dph6 mutants, which also lack the ability to form diphthamide but are resistant to growth inhibition by excess Dph5 levels. As judged from site-specific mutagenesis, the amidation reaction itself relies on a conserved ATP binding domain in Dph6 that, when altered, blocks diphthamide formation and confers resistance to eEF2 inhibition by sordarin.
Figures




References
-
- Collier R.J. Understanding the mode of action of diphtheria toxin: A perspective on progress during the 20th century. Toxicon. 2001;39:1793–1803. - PubMed
-
- Uthman S., Liu S., Giorgini F., Stark M.J.R., Costanzo M., Schaffrath R. Diphtheria Disease and Genes Involved in Formation of Diphthamide, Key Effector of the Diphtheria Toxin. In: Kumar R., editor. Insight and Control of Infectious Disease in Global Scenario. INTECH Open Access Publisher; Rijeka, Croatia: 2012. pp. 333–356.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

