Ubiquitin-specific proteases UBP12 and UBP13 act in circadian clock and photoperiodic flowering regulation in Arabidopsis
- PMID: 23645632
- PMCID: PMC3668078
- DOI: 10.1104/pp.112.213009
Ubiquitin-specific proteases UBP12 and UBP13 act in circadian clock and photoperiodic flowering regulation in Arabidopsis
Abstract
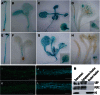
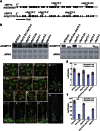
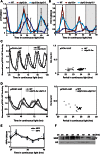
Protein ubiquitination is involved in most cellular processes. In Arabidopsis (Arabidopsis thaliana), ubiquitin-mediated protein degradation regulates the stability of key components of the circadian clock feedback loops and the photoperiodic flowering pathway. Here, we identified two ubiquitin-specific proteases, UBP12 and UBP13, involved in circadian clock and photoperiodic flowering regulation. Double mutants of ubp12 and ubp13 display pleiotropic phenotypes, including early flowering and short periodicity of circadian rhythms. In ubp12 ubp13 double mutants, CONSTANS (CO) transcript rises earlier than that of wild-type plants during the day, which leads to increased expression of FLOWERING LOCUS T. This, and analysis of ubp12 co mutants, indicates that UBP12 and UBP13 regulate photoperiodic flowering through a CO-dependent pathway. In addition, UBP12 and UBP13 regulate the circadian rhythm of clock genes, including LATE ELONGATED HYPOCOTYL, CIRCADIAN CLOCK ASSOCIATED1, and TIMING OF CAB EXPRESSION1. Furthermore, UBP12 and UBP13 are circadian controlled. Therefore, our work reveals a role for two deubiquitinases, UBP12 and UBP13, in the control of the circadian clock and photoperiodic flowering, which extends our understanding of ubiquitin in daylength measurement in higher plants.
Figures


Similar articles
-
LATE ELONGATED HYPOCOTYL regulates photoperiodic flowering via the circadian clock in Arabidopsis.BMC Plant Biol. 2016 May 20;16(1):114. doi: 10.1186/s12870-016-0810-8. BMC Plant Biol. 2016. PMID: 27207270 Free PMC article.
-
Two new clock proteins, LWD1 and LWD2, regulate Arabidopsis photoperiodic flowering.Plant Physiol. 2008 Oct;148(2):948-59. doi: 10.1104/pp.108.124917. Epub 2008 Aug 1. Plant Physiol. 2008. PMID: 18676661 Free PMC article.
-
GIGANTEA recruits the UBP12 and UBP13 deubiquitylases to regulate accumulation of the ZTL photoreceptor complex.Nat Commun. 2019 Aug 21;10(1):3750. doi: 10.1038/s41467-019-11769-7. Nat Commun. 2019. PMID: 31434902 Free PMC article.
-
Circadian clock and photoperiodic response in Arabidopsis: from seasonal flowering to redox homeostasis.Biochemistry. 2015 Jan 20;54(2):157-70. doi: 10.1021/bi500922q. Epub 2014 Dec 30. Biochemistry. 2015. PMID: 25346271 Free PMC article. Review.
-
Time to flower: interplay between photoperiod and the circadian clock.J Exp Bot. 2015 Feb;66(3):719-30. doi: 10.1093/jxb/eru441. Epub 2014 Nov 4. J Exp Bot. 2015. PMID: 25371508 Review.
Cited by
-
Lipid-mediated activation of plasma membrane-localized deubiquitylating enzymes modulate endosomal trafficking.Nat Commun. 2022 Nov 12;13(1):6897. doi: 10.1038/s41467-022-34637-3. Nat Commun. 2022. PMID: 36371501 Free PMC article.
-
Genome-wide identification and expression analysis of Ubiquitin-specific protease gene family in maize (Zea mays L.).BMC Plant Biol. 2024 May 16;24(1):404. doi: 10.1186/s12870-024-04953-5. BMC Plant Biol. 2024. PMID: 38750451 Free PMC article.
-
Chromatin sensing: integration of environmental signals to reprogram plant development through chromatin regulators.J Exp Bot. 2024 Jul 23;75(14):4332-4345. doi: 10.1093/jxb/erae086. J Exp Bot. 2024. PMID: 38436409 Free PMC article. Review.
-
The ubiquitin system affects agronomic plant traits.J Biol Chem. 2020 Oct 2;295(40):13940-13955. doi: 10.1074/jbc.REV120.011303. Epub 2020 Aug 12. J Biol Chem. 2020. PMID: 32796036 Free PMC article. Review.
-
Genome-wide detection of genotype environment interactions for flowering time in Brassica napus.Front Plant Sci. 2022 Nov 21;13:1065766. doi: 10.3389/fpls.2022.1065766. eCollection 2022. Front Plant Sci. 2022. PMID: 36479520 Free PMC article.
References
-
- Alabadí D, Oyama T, Yanovsky MJ, Harmon FG, Más P, Kay SA. (2001) Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293: 880–883 - PubMed
-
- Carré IA, Kim JY. (2002) MYB transcription factors in the Arabidopsis circadian clock. J Exp Bot 53: 1551–1557 - PubMed
-
- Chandler JS, McArdle B, Callis J. (1997) AtUBP3 and AtUBP4 are two closely related Arabidopsis thaliana ubiquitin-specific proteases present in the nucleus. Mol Gen Genet 255: 302–310 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

