RegaDB: community-driven data management and analysis for infectious diseases
- PMID: 23645815
- PMCID: PMC3661054
- DOI: 10.1093/bioinformatics/btt162
RegaDB: community-driven data management and analysis for infectious diseases
Abstract
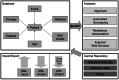
Summary: RegaDB is a free and open source data management and analysis environment for infectious diseases. RegaDB allows clinicians to store, manage and analyse patient data, including viral genetic sequences. Moreover, RegaDB provides researchers with a mechanism to collect data in a uniform format and offers them a canvas to make newly developed bioinformatics tools available to clinicians and virologists through a user friendly interface.
Availability and implementation: Source code, binaries and documentation are available on http://rega.kuleuven.be/cev/regadb. RegaDB is written in the Java programming language, using a web-service-oriented architecture.
Figures
References
-
- Altschul SF, et al. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. - PubMed
-
- Assel M, et al. A collaborative environment allowing clinical investigations on integrated biomedical databases. Stud. Health Technol. Inform. 2009;147:51–61. - PubMed
-
- de Oliveira T, et al. An integrated genetic data environment (GDE)-based Linux interface for analysis of HIV-1 and other microbial sequences. Bioinformatics. 2003;19:153–154. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical