Alternative excision repair pathways
- PMID: 23645854
- PMCID: PMC3660826
- DOI: 10.1101/cshperspect.a012617
Alternative excision repair pathways
Abstract
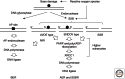
Alternative excision repair (AER) is a category of excision repair initiated by a single nick, made by an endonuclease, near the site of DNA damage, and followed by excision of the damaged DNA, repair synthesis, and ligation. The ultraviolet (UV) damage endonuclease in fungi and bacteria introduces a nick immediately 5' to various types of UV damage and initiates its excision repair that is independent of nucleotide excision repair (NER). Endo IV-type apurinic/apyrimidinic (AP) endonucleases from Escherichia coli and yeast and human Exo III-type AP endonuclease APEX1 introduce a nick directly and immediately 5' to various types of oxidative base damage besides the AP site, initiating excision repair. Another endonuclease, endonuclease V from bacteria to humans, binds deaminated bases and cleaves the phosphodiester bond located 1 nucleotide 3' of the base, leading to excision repair. A single-strand break in DNA is one of the most frequent types of DNA damage within cells and is repaired efficiently. AER makes use of such repair capability of single-strand breaks, removes DNA damage, and has an important role in complementing BER and NER.
Figures


Similar articles
-
The major Arabidopsis thaliana apurinic/apyrimidinic endonuclease, ARP is involved in the plant nucleotide incision repair pathway.DNA Repair (Amst). 2016 Dec;48:30-42. doi: 10.1016/j.dnarep.2016.10.009. Epub 2016 Oct 29. DNA Repair (Amst). 2016. PMID: 27836324
-
Abasic site recognition by two apurinic/apyrimidinic endonuclease families in DNA base excision repair: the 3' ends justify the means.Mutat Res. 2000 Aug 30;460(3-4):211-29. doi: 10.1016/s0921-8777(00)00028-8. Mutat Res. 2000. PMID: 10946230 Review.
-
Early steps in the DNA base excision/single-strand interruption repair pathway in mammalian cells.Cell Res. 2008 Jan;18(1):27-47. doi: 10.1038/cr.2008.8. Cell Res. 2008. PMID: 18166975 Free PMC article. Review.
-
Long patch base excision repair in mammalian mitochondrial genomes.J Biol Chem. 2008 Sep 26;283(39):26349-56. doi: 10.1074/jbc.M803491200. Epub 2008 Jul 17. J Biol Chem. 2008. PMID: 18635552 Free PMC article.
-
Repair of apurinic/apyrimidinic sites by UV damage endonuclease; a repair protein for UV and oxidative damage.Nucleic Acids Res. 1999 Aug 1;27(15):3096-103. doi: 10.1093/nar/27.15.3096. Nucleic Acids Res. 1999. PMID: 10454605 Free PMC article.
Cited by
-
Repair of Hypoxanthine in DNA Revealed by DNA Glycosylases and Endonucleases From Hyperthermophilic Archaea.Front Microbiol. 2021 Aug 31;12:736915. doi: 10.3389/fmicb.2021.736915. eCollection 2021. Front Microbiol. 2021. PMID: 34531846 Free PMC article. Review.
-
Structural basis for recognition of distinct deaminated DNA lesions by endonuclease Q.Proc Natl Acad Sci U S A. 2021 Mar 9;118(10):e2021120118. doi: 10.1073/pnas.2021120118. Proc Natl Acad Sci U S A. 2021. PMID: 33658373 Free PMC article.
-
DNA Polymerase B1 Binding Protein 1 Is Important for DNA Repair by Holoenzyme PolB1 in the Extremely Thermophilic Crenarchaeon Sulfolobus acidocaldarius.Microorganisms. 2021 Feb 20;9(2):439. doi: 10.3390/microorganisms9020439. Microorganisms. 2021. PMID: 33672533 Free PMC article.
-
DNA Damage and Deficiencies in the Mechanisms of Its Repair: Implications in the Pathogenesis of Systemic Lupus Erythematosus.J Immunol Res. 2018 Jul 12;2018:8214379. doi: 10.1155/2018/8214379. eCollection 2018. J Immunol Res. 2018. PMID: 30116756 Free PMC article. Review.
-
Molecular Basis of Substrate Recognition of Endonuclease Q from the Euryarchaeon Pyrococcus furiosus.J Bacteriol. 2020 Jan 2;202(2):e00542-19. doi: 10.1128/JB.00542-19. Print 2020 Jan 2. J Bacteriol. 2020. PMID: 31685534 Free PMC article.
References
-
- Alleva JL, Zuo S, Hurwitz J, Doetsch PW 2000. In vitro reconstitution of the Schizosaccharomyces pombe alternative excision repair pathway. Biochemistry 39: 2659–2666 - PubMed
-
- Garcin ED, Hosfield DJ, Desai SA, Haas BJ, Bjoras M, Cunningham RP, Tainer JA 2008. DNA apurinic-apyrimidinic site binding and excision by endonuclease IV. Nat Struct Mol Biol 15: 515–522 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
