Light-scattering-based analysis of biomolecular interactions
- PMID: 23646069
- PMCID: PMC3641300
- DOI: 10.1007/s12551-013-0107-1
Light-scattering-based analysis of biomolecular interactions
Abstract
While light scattering has long been applied to the analysis of biomolecular interactions, recent advances have extended the practical use of light scattering techniques to cover a rather broad range of phenomena. In this paper I review essential light scattering theory as applied to specific interactions under thermodynamically ideal conditions and present examples showing how light scattering elucidates the dynamic equilibrium and kinetic behavior of proteins and other biomacromolecules.
Keywords: Affinity; CG-DLS; CG-MALS; Interactions; Light scattering; Stoichiometry.
Figures
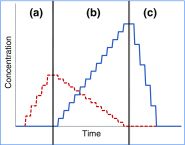
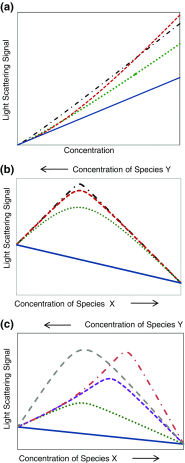
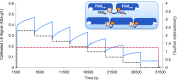
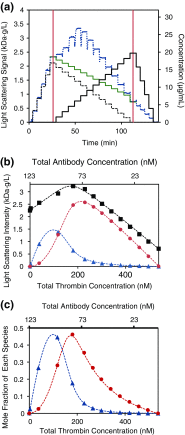
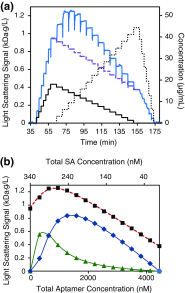
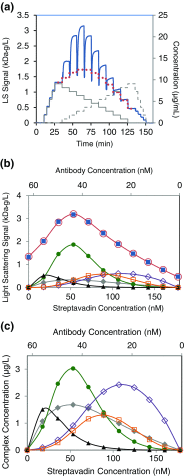
References
-
- Bajaj H, Sharma VK, Kalonia D. A high-throughput method for detection of protein self-association and second virial coefficient using size-exclusion chromatography through simultaneous measurement of concentration and scattered light intensity. Pharm Res. 2007;24:2071–2083. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

