Identification of ClpP substrates in Caulobacter crescentus reveals a role for regulated proteolysis in bacterial development
- PMID: 23647068
- PMCID: PMC3681837
- DOI: 10.1111/mmi.12241
Identification of ClpP substrates in Caulobacter crescentus reveals a role for regulated proteolysis in bacterial development
Abstract
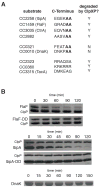
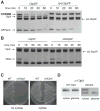
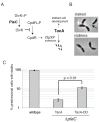
Energy-dependent proteases ensure the timely removal of unwanted proteins in a highly selective fashion. In Caulobacter crescentus, protein degradation by the ClpXP protease is critical for cell cycle progression; however, only a handful of substrates are currently known. Here, we use a trapping approach to identify putative substrates of the ClpP associated proteases in C. crescentus. Biochemical validation of several of these targets reveals specific protease recognition motifs and suggests a need for ClpXP-specific degradation beyond degradation of known cell cycle regulators. We focus on a particular instance of regulated proteolysis in Caulobacter by exploring the role of ClpXP in degrading the stalk synthesis transcription factor TacA. We show that TacA degradation is controlled during the cell cycle dependent on the ClpXP regulator CpdR and that stabilization of TacA increases degradation of another ClpXP substrate, CtrA, while restoring deficiencies associated with prolific CpdR activity. Together, our work reveals a number of new validated ClpXP substrates, clarifies rules of protease substrate selection, and demonstrates how regulated protein degradation is critical for Caulobacter development and cell cycle progression.
© 2013 John Wiley & Sons Ltd.
Figures


References
-
- Biondi EG, Reisinger SJ, Skerker JM, Arif M, Perchuk BS, Ryan KR, Laub MT. Regulation of the bacterial cell cycle by an integrated genetic circuit. Nature. 2006a;444:899–904. - PubMed
-
- Biondi EG, Skerker JM, Arif M, Prasol MS, Perchuk BS, Laub MT. A phosphorelay system controls stalk biogenesis during cell cycle progression in Caulobacter crescentus. Molecular microbiology. 2006b;59:386–401. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

