A roadmap of cell-type specific gene expression during sequential stages of the arbuscular mycorrhiza symbiosis
- PMID: 23647797
- PMCID: PMC3667144
- DOI: 10.1186/1471-2164-14-306
A roadmap of cell-type specific gene expression during sequential stages of the arbuscular mycorrhiza symbiosis
Abstract
Background: About 80% of today's land plants are able to establish an arbuscular mycorrhizal (AM) symbiosis with Glomeromycota fungi to improve their access to nutrients and water in the soil. On the molecular level, the development of AM symbioses is only partly understood, due to the asynchronous development of the microsymbionts in the host roots. Although many genes specifically activated during fungal colonization have been identified, genome-wide information on the exact place and time point of their activation remains limited.
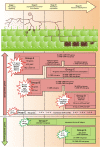
Results: In this study, we relied on a combination of laser-microdissection and the use of Medicago GeneChips to perform a genome-wide analysis of transcription patterns in defined cell-types of Medicago truncatula roots mycorrhized with Glomus intraradices. To cover major stages of AM development, we harvested cells at 5-6 and at 21 days post inoculation (dpi). Early developmental stages of the AM symbiosis were analysed by monitoring gene expression in appressorial and non-appressorial areas from roots harbouring infection units at 5-6 dpi. Here, the use of laser-microdissection for the first time enabled the targeted harvest of those sites, where fungal hyphae first penetrate the root. Circumventing contamination with developing arbuscules, we were able to specifically detect gene expression related to early infection events. To cover the late stages of AM formation, we studied arbusculated cells, cortical cells colonized by intraradical hyphae, and epidermal cells from mature mycorrhizal roots at 21 dpi. Taken together, the cell-specific expression patterns of 18014 genes were revealed, including 1392 genes whose transcription was influenced by mycorrhizal colonization at different stages, namely the pre-contact phase, the infection of roots via fungal appressoria, the subsequent colonization of the cortex by fungal hyphae, and finally the formation of arbuscules. Our cellular expression patterns identified distinct groups of AM-activated genes governing the sequential reprogramming of host roots towards an accommodation of microsymbionts, including 42 AM-activated transcription factor genes.
Conclusions: Our genome-wide analysis provides novel information on the cell-specific activity of AM-activated genes during both early and late stages of AM development, together revealing the road map of fine-tuned adjustments of transcript accumulation within root tissues during AM fungal colonization.
Figures





References
-
- Schüßler A, Schwarzott D, Walker C. A new fungal phylum, the Glomeromycota: phylogeny and evolution. Mycol Res. 2001;105:1413–1421. doi: 10.1017/S0953756201005196. - DOI
-
- Douds DD, Pfeffer PE, Shachar-Hill Y. In: Arbuscular Mycorrhizas Physiology and Function. Douds DD, Kalpunik Y, editor. Dordrecht, The Netherlands: Kluwer Academic Publishers; 2000. Carbon partioning, cost and metabolism of Arbuscular Mycorrhizae; pp. 107–130.
-
- Davies FT, Potter JR, Linderman RG. Drought Resistance of Mycorrhizal Pepper Plants Independent of Leaf P-Concentration - Response in Gas-Exchange and Water Relations. Physiol Plant. 1993;87(1):45–53. doi: 10.1111/j.1399-3054.1993.tb08789.x. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

