Genetic characterization of a core collection of flax (Linum usitatissimum L.) suitable for association mapping studies and evidence of divergent selection between fiber and linseed types
- PMID: 23647851
- PMCID: PMC3656786
- DOI: 10.1186/1471-2229-13-78
Genetic characterization of a core collection of flax (Linum usitatissimum L.) suitable for association mapping studies and evidence of divergent selection between fiber and linseed types
Abstract
Background: Flax is valued for its fiber, seed oil and nutraceuticals. Recently, the fiber industry has invested in the development of products made from linseed stems, making it a dual purpose crop. Simultaneous targeting of genomic regions controlling stem fiber and seed quality traits could enable the development of dual purpose cultivars. However, the genetic diversity, population structure and linkage disequilibrium (LD) patterns necessary for association mapping (AM) have not yet been assessed in flax because genomic resources have only recently been developed. We characterized 407 globally distributed flax accessions using 448 microsatellite markers. The data was analyzed to assess the suitability of this core collection for AM. Genomic scans to identify candidate genes selected during the divergent breeding process of fiber flax and linseed were conducted using the whole genome shotgun sequence of flax.
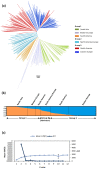
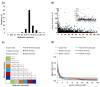
Results: Combined genetic structure analysis assigned all accessions to two major groups with six sub-groups. Population differentiation was weak between the major groups (F(ST) = 0.094) and for most of the pairwise comparisons among sub-groups. The molecular coancestry analysis indicated weak relatedness (mean = 0.287) for most individual pairs. Abundant genetic diversity was observed in the total panel (5.32 alleles per locus), and some sub-groups showed a high proportion of private alleles. The average genome-wide LD (r²) was 0.036, with a relatively fast decay of 1.5 cM. Genomic scans between fiber flax and linseed identified candidate genes involved in cell-wall biogenesis/modification, xylem identity and fatty acid biosynthesis congruent with genes previously identified in flax and other plant species.
Conclusions: Based on the abundant genetic diversity, weak population structure and relatedness and relatively fast LD decay, we concluded that this core collection is suitable for AM studies targeting multiple agronomic and quality traits aiming at the improvement of flax as a true dual purpose crop. Our genomic scans provide the first insights into candidate regions affected by divergent selection in flax. In combination with AM, genomic scans have the ability to increase the power to detect loci influencing complex traits.
Figures
Similar articles
-
Molecular Advances to Combat Different Biotic and Abiotic Stresses in Linseed (Linum usitatissimum L.): A Comprehensive Review.Genes (Basel). 2023 Jul 17;14(7):1461. doi: 10.3390/genes14071461. Genes (Basel). 2023. PMID: 37510365 Free PMC article. Review.
-
Association mapping of seed quality traits using the Canadian flax (Linum usitatissimum L.) core collection.Theor Appl Genet. 2014 Apr;127(4):881-96. doi: 10.1007/s00122-014-2264-4. Epub 2014 Jan 26. Theor Appl Genet. 2014. PMID: 24463785 Free PMC article.
-
Genomic variations and association study of agronomic traits in flax.BMC Genomics. 2018 Jul 3;19(1):512. doi: 10.1186/s12864-018-4899-z. BMC Genomics. 2018. PMID: 29969983 Free PMC article.
-
QTL for fatty acid composition and yield in linseed (Linum usitatissimum L.).Theor Appl Genet. 2015 May;128(5):965-84. doi: 10.1007/s00122-015-2483-3. Epub 2015 Mar 8. Theor Appl Genet. 2015. PMID: 25748113
-
History and prospects of flax genetic markers.Front Plant Sci. 2025 Jan 15;15:1495069. doi: 10.3389/fpls.2024.1495069. eCollection 2024. Front Plant Sci. 2025. PMID: 39881731 Free PMC article. Review.
Cited by
-
The Complex Genetic Architecture of Early Root and Shoot Traits in Flax Revealed by Genome-Wide Association Analyses.Front Plant Sci. 2019 Nov 19;10:1483. doi: 10.3389/fpls.2019.01483. eCollection 2019. Front Plant Sci. 2019. PMID: 31798617 Free PMC article.
-
Molecular Advances to Combat Different Biotic and Abiotic Stresses in Linseed (Linum usitatissimum L.): A Comprehensive Review.Genes (Basel). 2023 Jul 17;14(7):1461. doi: 10.3390/genes14071461. Genes (Basel). 2023. PMID: 37510365 Free PMC article. Review.
-
Analysis of genetic diversity and population structure of rice germplasm from north-eastern region of India and development of a core germplasm set.PLoS One. 2014 Nov 20;9(11):e113094. doi: 10.1371/journal.pone.0113094. eCollection 2014. PLoS One. 2014. PMID: 25412256 Free PMC article.
-
The potential of pale flax as a source of useful genetic variation for cultivated flax revealed through molecular diversity and association analyses.Mol Breed. 2014;34(4):2091-2107. doi: 10.1007/s11032-014-0165-5. Epub 2014 Aug 12. Mol Breed. 2014. PMID: 26316841 Free PMC article.
-
Genome-wide characterization of genetic diversity and population structure in Secale.BMC Plant Biol. 2014 Aug 1;14:184. doi: 10.1186/1471-2229-14-184. BMC Plant Biol. 2014. PMID: 25085433 Free PMC article.
References
-
- Vavilov NI. The origin, variation, immunity and breeding of cultivated plants. Chronica Botanica. 1951;13:1–366.
-
- Green AG, Chen Y, Singh SP, Dribnenki JCP. In: Compendium of transgenic crop plants. Kole C, Hall TC, editor. Oxford: Blackwell Publishing Ltd; 2008. Flax; pp. 199–226.
-
- Diederichsen A, Ulrich A. Variability in stem fibre content and its association with other characteristics in 1177 flax (Linum usitatissimum L) genebank accessions. Ind Crop Prod. 2009;30(1):33–39. doi: 10.1016/j.indcrop.2009.01.002. - DOI
-
- van Zeist W, Bakker-Heeres JAH. Evidence for linseed cultivation before 6000 BC. J Archeol Sci. 1975;2(3):215–219. doi: 10.1016/0305-4403(75)90059-X. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

