Characterization of a novel NKG2D and NKp46 double-mutant mouse reveals subtle variations in the NK cell repertoire
- PMID: 23649470
- PMCID: PMC3689249
- DOI: 10.1182/blood-2012-12-471607
Characterization of a novel NKG2D and NKp46 double-mutant mouse reveals subtle variations in the NK cell repertoire
Abstract
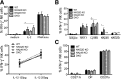
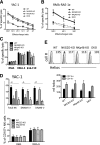
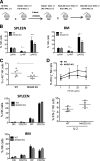
The immunoreceptors NKG2D and NKp46 are known for their capacity to activate natural killer (NK) cell cytotoxicity and secretory responses in the contexts of tumors and infections, yet their roles in NK cell education remain unclear. Here, we provide the first characterization of mice deficient for both NKG2D and NKp46 receptors to address the relevance of their concomitant absence during NK cell development and function. Our findings reveal that NK cells develop normally in double-mutant (DKO) mice. Mice lacking NKG2D but not NKp46 showed subtle differences in the percentages of NK cells expressing inhibitory Ly49 receptors and the adhesion molecule DNAM-1. A slightly increased percentage of terminally differentiated NK cells and functional response to in vitro stimuli was observed in some experiments. These alterations were modest and did not affect NK cell function in vivo in response to mouse cytomegalovirus infection. NKp46 deficiency alone, or in combination with NKG2D deficiency, had no effect on frequency or function of NK cells.
Figures





References
-
- Biassoni R, Cantoni C, Pende D, et al. Human natural killer cell receptors and co-receptors. Immunol Rev. 2001;181:203–214. - PubMed
-
- Lanier LL. NK cell recognition. Annu Rev Immunol. 2005;23:225–274. - PubMed
-
- Kim S, Iizuka K, Kang HS, et al. In vivo developmental stages in murine natural killer cell maturation. Nat Immunol. 2002;3(6):523–528. - PubMed
-
- Carotta S, Pang SH, Nutt SL, Belz GT. Identification of the earliest NK-cell precursor in the mouse BM. Blood. 2011;117(20):5449–5452. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

