Interferon-inducible transmembrane protein 3 (IFITM3) restricts reovirus cell entry
- PMID: 23649619
- PMCID: PMC3682530
- DOI: 10.1074/jbc.M112.438515
Interferon-inducible transmembrane protein 3 (IFITM3) restricts reovirus cell entry
Abstract
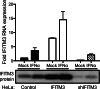
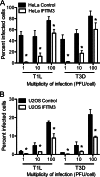
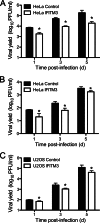
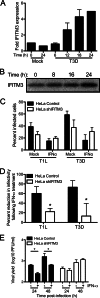
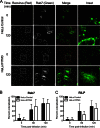
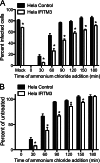
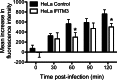
Reoviruses are double-stranded RNA viruses that infect the mammalian respiratory and gastrointestinal tract. Reovirus infection elicits production of type I interferons (IFNs), which trigger antiviral pathways through the induction of interferon-stimulated genes (ISGs). Although hundreds of ISGs have been identified, the functions of many of these genes are unknown. The interferon-inducible transmembrane (IFITM) proteins are one class of ISGs that restrict the cell entry of some enveloped viruses, including influenza A virus. One family member, IFITM3, localizes to late endosomes, where reoviruses undergo proteolytic disassembly; therefore, we sought to determine whether IFITM3 also restricts reovirus entry. IFITM3-expressing cell lines were less susceptible to infection by reovirus, as they exhibited significantly lower percentages of infected cells in comparison to control cells. Reovirus replication was also significantly reduced in IFITM3-expressing cells. Additionally, cells expressing an shRNA targeting IFITM3 exhibited a smaller decrease in infection after IFN treatment than the control cells, indicating that endogenous IFITM3 restricts reovirus infection. However, IFITM3 did not restrict entry of reovirus infectious subvirion particles (ISVPs), which do not require endosomal proteolysis, indicating that restriction occurs in the endocytic pathway. Proteolysis of outer capsid protein μ1 was delayed in IFITM3-expressing cells in comparison to control cells, suggesting that IFITM3 modulates the function of late endosomal compartments either by reducing the activity of endosomal proteases or delaying the proteolytic processing of virions. These data provide the first evidence that IFITM3 restricts infection by a nonenveloped virus and suggest that IFITM3 targets an increasing number of viruses through a shared requirement for endosomes during cell entry.
Keywords: Endosomes; Innate Immunity; Interferon; Interferon-stimulated Gene; Reovirus; Virus Entry.
Figures


References
-
- Borden E. C., Williams B. R. (2011) Interferon-stimulated genes and their protein products. what and how? J. Interferon Cytokine Res. 31, 1–4 - PubMed
-
- Farrell P. J., Balkow K., Hunt T., Jackson R. J., Trachsel H. (1977) Phosphorylation of initiation factor eIF-2 and the control of reticulocyte protein synthesis. Cell 11, 187–200 - PubMed
-
- Ratner L., Wiegand R. C., Farrell P. J., Sen G. C., Cabrer B., Lengyel P. (1978) Interferon, double-stranded RNA, and RNA degradation. Fractionation of the endonuclease INT system into two macromolecular components. Role of a small molecule in nuclease activation. Biochem. Biophys. Res. Commun. 81, 947–954 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

