HMGB1 protein does not mediate the inflammatory response in spontaneous spinal cord regeneration: a hint for CNS regeneration
- PMID: 23649623
- PMCID: PMC3689963
- DOI: 10.1074/jbc.M113.463810
HMGB1 protein does not mediate the inflammatory response in spontaneous spinal cord regeneration: a hint for CNS regeneration
Abstract
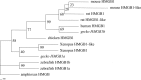
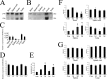
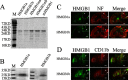
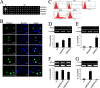
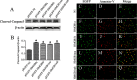
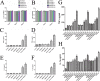
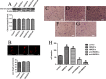
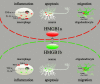
Uncontrolled, excessive inflammation contributes to the secondary tissue damage of traumatic spinal cord, and HMGB1 is highlighted for initiation of a vicious self-propagating inflammatory circle by release from necrotic cells or immune cells. Several regenerative-competent vertebrates have evolved to circumvent the second damages during the spontaneous spinal cord regeneration with an unknown HMGB1 regulatory mechanism. By genomic surveys, we have revealed that two paralogs of HMGB1 are broadly retained from fish in the phylogeny. However, their spatial-temporal expression and effects, as shown in lowest amniote gecko, were tightly controlled in order that limited inflammation was produced in spontaneous regeneration. Two paralogs from gecko HMGB1 (gHMGB1) yielded distinct injury and infectious responses, with gHMGB1b significantly up-regulated in the injured cord. The intracellular gHMGB1b induced less release of inflammatory cytokines than gHMGB1a in macrophages, and the effects could be shifted by exchanging one amino acid in the inflammatory domain. Both intracellular proteins were able to mediate neuronal programmed apoptosis, which has been indicated to produce negligible inflammatory responses. In vivo studies demonstrated that the extracellular proteins could not trigger a cascade of the inflammatory cytokines in the injured spinal cord. Signal transduction analysis found that gHMGB1 proteins could not bind with cell surface receptors TLR2 and TLR4 to activate inflammatory signaling pathway. However, they were able to interact with the receptor for advanced glycation end products to potentiate oligodendrocyte migration by activation of both NFκB and Rac1/Cdc42 signaling. Our results reveal that HMGB1 does not mediate the inflammatory response in spontaneous spinal cord regeneration, but it promotes CNS regeneration.
Keywords: CNS; Evolution; HMGB1; Inflammation; Receptor for Advanced Glycation End Products (RAGE); Regeneration; Toll-like receptors (TLR).
Figures


Similar articles
-
High mobility group box-1 (HMGB1) is increased in injured mouse spinal cord and can elicit neurotoxic inflammation.Brain Behav Immun. 2018 Aug;72:22-33. doi: 10.1016/j.bbi.2017.11.018. Epub 2017 Nov 23. Brain Behav Immun. 2018. PMID: 29175543 Free PMC article.
-
Toll-like receptor signaling in amyotrophic lateral sclerosis spinal cord tissue.Neuroscience. 2011 Apr 14;179:233-43. doi: 10.1016/j.neuroscience.2011.02.001. Epub 2011 Feb 12. Neuroscience. 2011. PMID: 21303685
-
Inhibiting HMGB1-RAGE axis prevents pro-inflammatory macrophages/microglia polarization and affords neuroprotection after spinal cord injury.J Neuroinflammation. 2020 Oct 9;17(1):295. doi: 10.1186/s12974-020-01973-4. J Neuroinflammation. 2020. PMID: 33036632 Free PMC article.
-
Expatiating the molecular approaches of HMGB1 in diabetes mellitus: Highlighting signalling pathways via RAGE and TLRs.Mol Biol Rep. 2021 Feb;48(2):1869-1881. doi: 10.1007/s11033-020-06130-x. Epub 2021 Jan 21. Mol Biol Rep. 2021. PMID: 33479829 Review.
-
Review: The role of HMGB1 in spinal cord injury.Front Immunol. 2023 Jan 12;13:1094925. doi: 10.3389/fimmu.2022.1094925. eCollection 2022. Front Immunol. 2023. PMID: 36713448 Free PMC article. Review.
Cited by
-
HMGB1/Advanced Glycation End Products (RAGE) does not aggravate inflammation but promote endogenous neural stem cells differentiation in spinal cord injury.Sci Rep. 2017 Sep 4;7(1):10332. doi: 10.1038/s41598-017-10611-8. Sci Rep. 2017. PMID: 28871209 Free PMC article.
-
High-mobility group box-1 as an autocrine trophic factor in white matter stroke.Proc Natl Acad Sci U S A. 2017 Jun 20;114(25):E4987-E4995. doi: 10.1073/pnas.1702035114. Epub 2017 Jun 5. Proc Natl Acad Sci U S A. 2017. PMID: 28584116 Free PMC article.
-
Adult astrocytes from reptiles are resistant to proinflammatory activation via sustaining Vav1 expression.J Biol Chem. 2021 Jan-Jun;296:100527. doi: 10.1016/j.jbc.2021.100527. Epub 2021 Mar 9. J Biol Chem. 2021. PMID: 33705794 Free PMC article.
-
Receptor for Advanced Glycation End-Products (RAGE) Blockade Do Damage to Neuronal Survival via Disrupting Wnt/β-Catenin Signaling in Spinal Cord Injury.Neurochem Res. 2018 Jul;43(7):1405-1412. doi: 10.1007/s11064-018-2555-2. Epub 2018 May 22. Neurochem Res. 2018. PMID: 29790067
-
High mobility group box 1 in the central nervous system: regeneration hidden beneath inflammation.Neural Regen Res. 2025 Jan 1;20(1):107-115. doi: 10.4103/NRR.NRR-D-23-01964. Epub 2024 Apr 3. Neural Regen Res. 2025. PMID: 38767480 Free PMC article.
References
-
- Popovich P. G., Longbrake E. E. (2008) Can the immune system be harnessed to repair the CNS? Nat. Rev. Neurosci. 9, 481–493 - PubMed
-
- Hausmann O. N. (2003) Post-traumatic inflammation following spinal cord injury. Spinal Cord 41, 369–378 - PubMed
-
- Horner P. J., Gage F. H. (2000) Regenerating the damaged central nervous system. Nature 407, 963–970 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

