X-ray structure of an AdoMet radical activase reveals an anaerobic solution for formylglycine posttranslational modification
- PMID: 23650368
- PMCID: PMC3666706
- DOI: 10.1073/pnas.1302417110
X-ray structure of an AdoMet radical activase reveals an anaerobic solution for formylglycine posttranslational modification
Abstract
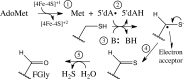
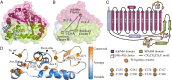
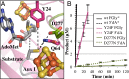
Arylsulfatases require a maturating enzyme to perform a co- or posttranslational modification to form a catalytically essential formylglycine (FGly) residue. In organisms that live aerobically, molecular oxygen is used enzymatically to oxidize cysteine to FGly. Under anaerobic conditions, S-adenosylmethionine (AdoMet) radical chemistry is used. Here we present the structures of an anaerobic sulfatase maturating enzyme (anSME), both with and without peptidyl-substrates, at 1.6-1.8 Å resolution. We find that anSMEs differ from their aerobic counterparts in using backbone-based hydrogen-bonding patterns to interact with their peptidyl-substrates, leading to decreased sequence specificity. These anSME structures from Clostridium perfringens are also the first of an AdoMet radical enzyme that performs dehydrogenase chemistry. Together with accompanying mutagenesis data, a mechanistic proposal is put forth for how AdoMet radical chemistry is coopted to perform a dehydrogenation reaction. In the oxidation of cysteine or serine to FGly by anSME, we identify D277 and an auxiliary [4Fe-4S] cluster as the likely acceptor of the final proton and electron, respectively. D277 and both auxiliary clusters are housed in a cysteine-rich C-terminal domain, termed SPASM domain, that contains homology to ~1,400 other unique AdoMet radical enzymes proposed to use [4Fe-4S] clusters to ligate peptidyl-substrates for subsequent modification. In contrast to this proposal, we find that neither auxiliary cluster in anSME bind substrate, and both are fully ligated by cysteine residues. Instead, our structural data suggest that the placement of these auxiliary clusters creates a conduit for electrons to travel from the buried substrate to the protein surface.
Keywords: iron–sulfur cluster fold; radical SAM dehydrogenase.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Schmidt B, Selmer T, Ingendoh A, von Figura K. A novel amino acid modification in sulfatases that is defective in multiple sulfatase deficiency. Cell. 1995;82(2):271–278. - PubMed
-
- Bojarová P, Williams SJ. Sulfotransferases, sulfatases and formylglycine-generating enzymes: A sulfation fascination. Curr Opin Chem Biol. 2008;12(5):573–581. - PubMed
-
- Dierks T, et al. Multiple sulfatase deficiency is caused by mutations in the gene encoding the human C(alpha)-formylglycine generating enzyme. Cell. 2003;113(4):435–444. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

