Mesenchymal glioma stem cells are maintained by activated glycolytic metabolism involving aldehyde dehydrogenase 1A3
- PMID: 23650391
- PMCID: PMC3666732
- DOI: 10.1073/pnas.1221478110
Mesenchymal glioma stem cells are maintained by activated glycolytic metabolism involving aldehyde dehydrogenase 1A3
Abstract
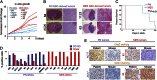
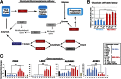
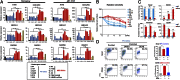
Tumor heterogeneity of high-grade glioma (HGG) is recognized by four clinically relevant subtypes based on core gene signatures. However, molecular signaling in glioma stem cells (GSCs) in individual HGG subtypes is poorly characterized. Here we identified and characterized two mutually exclusive GSC subtypes with distinct dysregulated signaling pathways. Analysis of mRNA profiles distinguished proneural (PN) from mesenchymal (Mes) GSCs and revealed a pronounced correlation with the corresponding PN or Mes HGGs. Mes GSCs displayed more aggressive phenotypes in vitro and as intracranial xenografts in mice. Further, Mes GSCs were markedly resistant to radiation compared with PN GSCs. The glycolytic pathway, comprising aldehyde dehydrogenase (ALDH) family genes and in particular ALDH1A3, were enriched in Mes GSCs. Glycolytic activity and ALDH activity were significantly elevated in Mes GSCs but not in PN GSCs. Expression of ALDH1A3 was also increased in clinical HGG compared with low-grade glioma or normal brain tissue. Moreover, inhibition of ALDH1A3 attenuated the growth of Mes but not PN GSCs. Last, radiation treatment of PN GSCs up-regulated Mes-associated markers and down-regulated PN-associated markers, whereas inhibition of ALDH1A3 attenuated an irradiation-induced gain of Mes identity in PN GSCs. Taken together, our data suggest that two subtypes of GSCs, harboring distinct metabolic signaling pathways, represent intertumoral glioma heterogeneity and highlight previously unidentified roles of ALDH1A3-associated signaling that promotes aberrant proliferation of Mes HGGs and GSCs. Inhibition of ALDH1A3-mediated pathways therefore might provide a promising therapeutic approach for a subset of HGGs with the Mes signature.
Keywords: cancer stem cell; epithelial-to-mesenchymal transition; glioblastoma; glioblastoma multiforme; proneural-to-mesenchymal transition.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Adamson C, et al. Glioblastoma multiforme: A review of where we have been and where we are going. Expert Opin Investig Drugs. 2009;18(8):1061–1083. - PubMed
-
- Phillips HS, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9(3):157–173. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R44 GM087798/GM/NIGMS NIH HHS/United States
- CA158911/CA/NCI NIH HHS/United States
- LM009657/LM/NLM NIH HHS/United States
- UL1 RR024153/RR/NCRR NIH HHS/United States
- P30 CA047904/CA/NCI NIH HHS/United States
- R01 CA130966/CA/NCI NIH HHS/United States
- NS037704/NS/NINDS NIH HHS/United States
- GM099213/GM/NIGMS NIH HHS/United States
- R43 GM087798/GM/NIGMS NIH HHS/United States
- CA130966/CA/NCI NIH HHS/United States
- CA135013/CA/NCI NIH HHS/United States
- R21 CA135013/CA/NCI NIH HHS/United States
- R01 CA148629/CA/NCI NIH HHS/United States
- CA148629/CA/NCI NIH HHS/United States
- R01 NS037704/NS/NINDS NIH HHS/United States
- ES019498/ES/NIEHS NIH HHS/United States
- GM087798/GM/NIGMS NIH HHS/United States
- UL1RR024153/RR/NCRR NIH HHS/United States
- R43 GM099213/GM/NIGMS NIH HHS/United States
- R21 ES019498/ES/NIEHS NIH HHS/United States
- R01 LM009657/LM/NLM NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

