Measuring EGFR separations on cells with ~10 nm resolution via fluorophore localization imaging with photobleaching
- PMID: 23650512
- PMCID: PMC3641073
- DOI: 10.1371/journal.pone.0062331
Measuring EGFR separations on cells with ~10 nm resolution via fluorophore localization imaging with photobleaching
Abstract
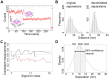
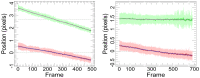
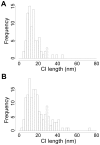
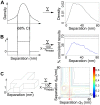
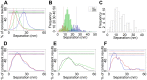
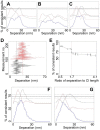
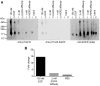
Detecting receptor dimerisation and other forms of clustering on the cell surface depends on methods capable of determining protein-protein separations with high resolution in the ~10-50 nm range. However, this distance range poses a significant challenge because it is too large for fluorescence resonance energy transfer and contains distances too small for all other techniques capable of high-resolution in cells. Here we have adapted the technique of fluorophore localisation imaging with photobleaching to measure inter-receptor separations in the cellular environment. Using the epidermal growth factor receptor, a key cancer target molecule, we demonstrate ~10 nm resolution while continuously covering the range of ~10-80 nm. By labelling the receptor on cells expressing low receptor numbers with a fluorescent antagonist we have found inter-receptor separations all the way up from 8 nm to 59 nm. Our data are consistent with epidermal growth factor receptors being able to form homo-polymers of at least 10 receptors in the absence of activating ligands.
Conflict of interest statement
Figures



References
-
- Ferguson KM, Berger MB, Mendrola JM, Cho HS, Leahy DJ, et al. (2003) EGF activates its receptor by removing interactions that autoinhibit ectodomain dimerization. Mol Cell 11: 507–517. - PubMed
-
- Garrett TP, McKern NM, Lou M, Elleman TC, Adams TE, et al. (2002) Crystal structure of a truncated epidermal growth factor receptor extracellular domain bound to transforming growth factor alpha. Cell 110: 763–773. - PubMed
-
- Ogiso H, Ishitani R, Nureki O, Fukai S, Yamanaka M, et al. (2002) Crystal structure of the complex of human epidermal growth factor and receptor extracellular domains. Cell 110: 775–787. - PubMed
-
- Zhang X, Gureasko J, Shen K, Cole PA, Kuriyan J (2006) An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell 125: 1137–1149. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

