SV40 late protein VP4 forms toroidal pores to disrupt membranes for viral release
- PMID: 23651212
- PMCID: PMC4235334
- DOI: 10.1021/bi400036z
SV40 late protein VP4 forms toroidal pores to disrupt membranes for viral release
Abstract
Nonenveloped viruses are generally released from the cell by the timely lysis of host cell membranes. SV40 has been used as a model virus for the study of the lytic nonenveloped virus life cycle. The expression of SV40 VP4 at later times during infection is concomitant with cell lysis. To investigate the role of VP4 in viral release and its mechanism of action, VP4 was expressed and purified from bacteria as a fusion protein for use in membrane disruption assays. Purified VP4 perforated membranes as demonstrated by the release of fluorescent markers encapsulated within large unilamellar vesicles or liposomes. Dynamic light scattering results revealed that VP4 treatment did not cause membrane lysis or change the size of the liposomes. Liposomes encapsulated with 4,4-difluoro-5,7-dimethyl-4-bora-3a,4a-diaza-3-indacene-labeled streptavidin were used to show that VP4 formed stable pores in membranes. These VP4 pores had an inner diameter of 1-5 nm. Asymmetrical liposomes containing pyrene-labeled lipids in the outer monolayer were employed to monitor transbilayer lipid diffusion. Consistent with VP4 forming toroidal pore structures in membranes, VP4 induced transbilayer lipid diffusion or lipid flip-flop. Altogether, these studies support a central role for VP4 acting as a viroporin in the disruption of cellular membranes to trigger SV40 viral release by forming toroidal pores that unite the outer and inner leaflets of membrane bilayers.
Figures



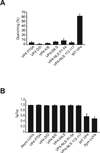
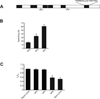
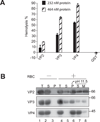
References
-
- Bieniasz PD. Late budding domains and host proteins in enveloped virus release. Virology. 2006;344:55–63. - PubMed
-
- Aldabe R, Barco A, Carrasco L. Membrane permeabilization by poliovirus proteins 2B and 2BC. J Biol Chem. 1996;271:23134–23137. - PubMed
-
- Han Z, Harty RN. The NS3 protein of bluetongue virus exhibits viroporin-like properties. J Biol Chem. 2004;279:43092–43097. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

