Direct evidence for RNA-RNA interactions at the 3' end of the Hepatitis C virus genome using surface plasmon resonance
- PMID: 23651615
- PMCID: PMC3683932
- DOI: 10.1261/rna.037606.112
Direct evidence for RNA-RNA interactions at the 3' end of the Hepatitis C virus genome using surface plasmon resonance
Abstract
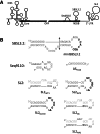
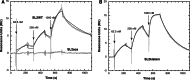
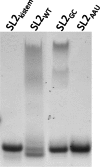
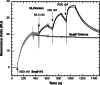
Surface plasmon resonance was used to investigate two previously described interactions analyzed by reverse genetics and complementation mutation experiments, involving 5BSL3.2, a stem-loop located in the NS5B coding region of HCV. 5BSL3.2 was immobilized on a sensor chip by streptavidin-biotin coupling, and its interaction either with the SL2 stem-loop of the 3' end or with an upstream sequence centered on nucleotide 9110 (referred to as Seq9110) was monitored in real-time. In contrast with previous results obtained by NMR assays with the same short RNA sequences that we used or SHAPE analysis with longer RNAs, we demonstrate that recognition between 5BSL3.2 and SL2 can occur in solution through a kissing-loop interaction. We show that recognition between Seq9110 and the internal loop of 5BSL3.2 does not prevent binding of SL2 on the apical loop of 5BSL3.2 and does not influence the rate constants of the SL2-5BSL3.2 complex. Therefore, the two binding sites of 5BSL3.2, the apical and internal loops, are structurally independent and both interactions can coexist. We finally show that the stem-loop SL2 is a highly dynamic RNA motif that fluctuates between at least two conformations: One is able to hybridize with 5BSL3.2 through loop-loop interaction, and the other one is capable of self-associating in the absence of protein, reinforcing the hypothesis of SL2 being a dimerization sequence. This result suggests also that the conformational dynamics of SL2 could play a crucial role for controlling the destiny of the genomic RNA.
Keywords: Hepatitis C virus; RNA; SPR; interactions; kinetics; kissing loop.
Figures

Similar articles
-
An unexpected RNA distal interaction mode found in an essential region of the hepatitis C virus genome.Nucleic Acids Res. 2018 May 4;46(8):4200-4212. doi: 10.1093/nar/gky074. Nucleic Acids Res. 2018. PMID: 29409065 Free PMC article.
-
3' RNA elements in hepatitis C virus replication: kissing partners and long poly(U).J Virol. 2008 Jan;82(1):184-95. doi: 10.1128/JVI.01796-07. Epub 2007 Oct 17. J Virol. 2008. PMID: 17942554 Free PMC article.
-
Mutations of the SL2 dimerization sequence of the hepatitis C genome abrogate viral replication.Cell Mol Life Sci. 2015 Sep;72(17):3375-85. doi: 10.1007/s00018-015-1893-3. Epub 2015 Mar 28. Cell Mol Life Sci. 2015. PMID: 25822205 Free PMC article.
-
The 5BSL3.2 Functional RNA Domain Connects Distant Regions in the Hepatitis C Virus Genome.Front Microbiol. 2017 Oct 31;8:2093. doi: 10.3389/fmicb.2017.02093. eCollection 2017. Front Microbiol. 2017. PMID: 29163393 Free PMC article. Review.
-
RNA structural elements of hepatitis C virus controlling viral RNA translation and the implications for viral pathogenesis.Viruses. 2012 Oct 19;4(10):2233-50. doi: 10.3390/v4102233. Viruses. 2012. PMID: 23202462 Free PMC article. Review.
Cited by
-
Inhibition of HCV translation by disrupting the structure and interactions of the viral CRE and 3' X-tail.Nucleic Acids Res. 2015 Mar 11;43(5):2914-26. doi: 10.1093/nar/gkv142. Epub 2015 Feb 20. Nucleic Acids Res. 2015. PMID: 25712095 Free PMC article.
-
Three-dimensional structure of the 3'X-tail of hepatitis C virus RNA in monomeric and dimeric states.RNA. 2017 Sep;23(9):1465-1476. doi: 10.1261/rna.060632.117. Epub 2017 Jun 19. RNA. 2017. PMID: 28630140 Free PMC article.
-
Nuclear Magnetic Resonance Study of RNA Structures at the 3'-End of the Hepatitis C Virus Genome.Biochemistry. 2017 Sep 19;56(37):4972-4984. doi: 10.1021/acs.biochem.7b00573. Epub 2017 Sep 6. Biochemistry. 2017. PMID: 28829576 Free PMC article.
-
A comprehensive review on plasmonic-based biosensors used in viral diagnostics.Commun Biol. 2021 Jan 15;4(1):70. doi: 10.1038/s42003-020-01615-8. Commun Biol. 2021. PMID: 33452375 Free PMC article. Review.
-
Shape matters: size-exclusion HPLC for the study of nucleic acid structural polymorphism.Nucleic Acids Res. 2014 Oct 29;42(19):e149. doi: 10.1093/nar/gku751. Epub 2014 Aug 20. Nucleic Acids Res. 2014. PMID: 25143531 Free PMC article.
References
-
- Aldaz-Carroll L, Tallet B, Dausse E, Yurchenko L, Toulmé JJ 2002. Apical loop–internal loop interactions: A new RNA–RNA recognition motif identified through in vitro selection against RNA hairpins of the hepatitis C virus mRNA. Biochemistry 41: 5883–5893 - PubMed
-
- Chang KY, Tinoco I Jr 1997. The structure of an RNA “kissing” hairpin complex of the HIV TAR hairpin loop and its complement. J Mol Biol 269: 52–66 - PubMed
-
- Christopeit T, Gossas T, Danielson UH 2009. Characterization of Ca2+ and phosphocholine interactions with C-reactive protein using a surface plasmon resonance biosensor. Anal Biochem 391: 39–44 - PubMed
-
- Clarke B 1997. Molecular virology of hepatitis C virus. J Gen Virol 78: 2397–2410 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
