The DEAH-box helicase DHX36 mediates dendritic localization of the neuronal precursor-microRNA-134
- PMID: 23651854
- PMCID: PMC3656329
- DOI: 10.1101/gad.211243.112
The DEAH-box helicase DHX36 mediates dendritic localization of the neuronal precursor-microRNA-134
Erratum in
- Genes Dev. 2013 Jul 15;27(14):1633
Abstract
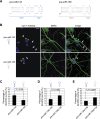
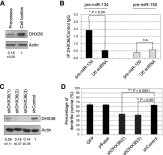
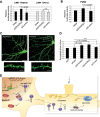
Specific microRNAs (miRNAs), including miR-134, localize to neuronal dendrites, where they control synaptic protein synthesis and plasticity. However, the mechanism of miRNA transport is unknown. We found that the neuronal precursor-miRNA-134 (pre-miR-134) accumulates in dendrites of hippocampal neurons and at synapses in vivo. Dendritic localization of pre-miR-134 is mediated by the DEAH-box helicase DHX36, which directly associates with the pre-miR-134 terminal loop. DHX36 function is required for miR-134-dependent inhibition of target gene expression and the control of dendritic spine size. Dendritically localized pre-miR-134 could provide a local source of miR-134 that can be mobilized in an activity-dependent manner during plasticity.
Keywords: DHX36; dendritic transport; precursor-microRNA; protein synthesis; synaptic plasticity.
Figures


References
-
- Ashraf SI, McLoon AL, Sclarsic SM, Kunes S 2006. Synaptic protein synthesis associated with memory is regulated by the RISC pathway in Drosophila. Cell 124: 191–205 - PubMed
-
- Banerjee S, Neveu P, Kosik KS 2009. A coordinated local translational control point at the synapse involving relief from silencing and MOV10 degradation. Neuron 64: 871–884 - PubMed
-
- Bramham CR, Wells DG 2007. Dendritic mRNA: Transport, translation and function. Nat Rev Neurosci 8: 776–789 - PubMed
-
- Brosius J, Tiedge H 2001. Neuronal BC1 RNA: Intracellular transport and activity-dependent modulation. Results Probl Cell Differ 34: 129–138 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
