GANP regulates recruitment of AID to immunoglobulin variable regions by modulating transcription and nucleosome occupancy
- PMID: 23652018
- PMCID: PMC3674236
- DOI: 10.1038/ncomms2823
GANP regulates recruitment of AID to immunoglobulin variable regions by modulating transcription and nucleosome occupancy
Abstract
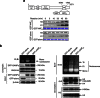
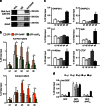
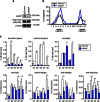
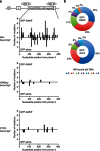
Somatic hypermutation in B cells is initiated by activation-induced cytidine deaminase-catalyzed C→U deamination at immunoglobulin variable regions. Here we investigate the role of the germinal centre-associated nuclear protein (GANP) in enhancing the access of activation-induced cytidine deaminase (AID) to immunoglobulin variable regions. We show that the nuclear export factor GANP is involved in chromatin modification at rearranged immunoglobulin variable loci, and its activity requires a histone acetyltransferase domain. GANP interacts with the transcription stalling protein Spt5 and facilitates RNA Pol-II recruitment to immunoglobulin variable regions. Germinal centre B cells from ganp-transgenic mice showed a higher AID occupancy at the immunoglobulin variable region, whereas B cells from conditional ganp-knockout mice exhibit a lower AID accessibility. These findings suggest that GANP-mediated chromatin modification promotes transcription complex recruitment and positioning at immunoglobulin variable loci to favour AID targeting.
Figures


Similar articles
-
GANP-mediated recruitment of activation-induced cytidine deaminase to cell nuclei and to immunoglobulin variable region DNA.J Biol Chem. 2010 Jul 30;285(31):23945-53. doi: 10.1074/jbc.M110.131441. Epub 2010 May 27. J Biol Chem. 2010. PMID: 20507984 Free PMC article.
-
Integrity of immunoglobulin variable regions is supported by GANP during AID-induced somatic hypermutation in germinal center B cells.Int Immunol. 2017 May 1;29(5):211-220. doi: 10.1093/intimm/dxx032. Int Immunol. 2017. PMID: 28541550 Free PMC article.
-
Germinal Center B-Cell-Associated Nuclear Protein (GANP) Involved in RNA Metabolism for B Cell Maturation.Adv Immunol. 2016;131:135-86. doi: 10.1016/bs.ai.2016.02.003. Epub 2016 Mar 29. Adv Immunol. 2016. PMID: 27235683 Review.
-
GANP regulates the choice of DNA repair pathway by DNA-PKcs interaction in AID-dependent IgV region diversification.J Immunol. 2014 Jun 15;192(12):5529-39. doi: 10.4049/jimmunol.1400021. Epub 2014 May 7. J Immunol. 2014. PMID: 24808370
-
Molecular mechanism of immunoglobulin V-region diversification regulated by transcription and RNA metabolism in antigen-driven B cells.Scand J Immunol. 2011 Jun;73(6):520-6. doi: 10.1111/j.1365-3083.2011.02557.x. Scand J Immunol. 2011. PMID: 21388430 Review.
Cited by
-
Sonic Hedgehog Pathway Modulation Normalizes Expression of Olig2 in Rostrally Patterned NPCs With Trisomy 21.Front Cell Neurosci. 2022 Jan 4;15:794675. doi: 10.3389/fncel.2021.794675. eCollection 2021. Front Cell Neurosci. 2022. PMID: 35058753 Free PMC article.
-
Development of a single-chain variable antibody fragment against a conserved region of the SARS-CoV-2 spike protein.Sci Rep. 2024 Jun 22;14(1):14419. doi: 10.1038/s41598-024-64103-7. Sci Rep. 2024. PMID: 38909102 Free PMC article.
-
A double-strand break can trigger immunoglobulin gene conversion.Nucleic Acids Res. 2017 Jan 9;45(1):231-243. doi: 10.1093/nar/gkw887. Epub 2016 Oct 3. Nucleic Acids Res. 2017. PMID: 27701075 Free PMC article.
-
Lysine Acetylation Goes Global: From Epigenetics to Metabolism and Therapeutics.Chem Rev. 2018 Feb 14;118(3):1216-1252. doi: 10.1021/acs.chemrev.7b00181. Epub 2018 Feb 6. Chem Rev. 2018. PMID: 29405707 Free PMC article. Review.
-
Humanized Immunoglobulin Mice: Models for HIV Vaccine Testing and Studying the Broadly Neutralizing Antibody Problem.Adv Immunol. 2017;134:235-352. doi: 10.1016/bs.ai.2017.01.004. Adv Immunol. 2017. PMID: 28413022 Free PMC article. Review.
References
-
- Peled J. U. et al.. The biochemistry of somatic hypermutation. Annu. Rev. Immunol. 26, 481–511 (2008) . - PubMed
-
- Odegard V. H. & Schatz D. G.. Targeting of somatic hypermutation. Nat. Rev. Immunol. 6, 573–583 (2006) . - PubMed
-
- Di Noia J. M. & Neuberger M. S. Molecular mechanisms of antibody somatic hypermutation. Annu. Rev. Biochem. 76, 1–22 (2007) . - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

