Inferring phylogeny and introgression using RADseq data: an example from flowering plants (Pedicularis: Orobanchaceae)
- PMID: 23652346
- PMCID: PMC3739883
- DOI: 10.1093/sysbio/syt032
Inferring phylogeny and introgression using RADseq data: an example from flowering plants (Pedicularis: Orobanchaceae)
Abstract
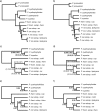
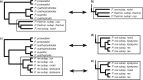
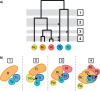
Phylogenetic relationships among recently diverged species are often difficult to resolve due to insufficient phylogenetic signal in available markers and/or conflict among gene trees. Here we explore the use of reduced-representation genome sequencing, specifically in the form of restriction-site associated DNA (RAD), for phylogenetic inference and the detection of ancestral hybridization in non-model organisms. As a case study, we investigate Pedicularis section Cyathophora, a systematically recalcitrant clade of flowering plants in the broomrape family (Orobanchaceae). Two methods of phylogenetic inference, maximum likelihood and Bayesian concordance, were applied to data sets that included as many as 40,000 RAD loci. Both methods yielded similar topologies that included two major clades: a "rex-thamnophila" clade, composed of two species and several subspecies with relatively low floral diversity, and geographically widespread distributions at lower elevations, and a "superba" clade, composed of three species characterized by relatively high floral diversity and isolated geographic distributions at higher elevations. Levels of molecular divergence between subspecies in the rex-thamnophila clade are similar to those between species in the superba clade. Using Patterson's D-statistic test, including a novel extension of the method that enables finer-grained resolution of introgression among multiple candidate taxa by removing the effect of their shared ancestry, we detect significant introgression among nearly all taxa in the rex-thamnophila clade, but not between clades or among taxa within the superba clade. These results suggest an important role for geographic isolation in the emergence of species barriers, by facilitating local adaptation and differentiation in the absence of homogenizing gene flow.
Figures



References
-
- Ané C. Reconstructing concordance trees and testing the coalescent model from genome-wide data sets. In: Knowles L.L., Kubatko L.S., editors. Estimating species trees: practical and theoretical aspects. Hoboken, NJ: John Wiley & Sons; 2010. pp. 35–51.
-
- Ané C., Larget B., Baum D.A., Smith S.D., Rokas A. Bayesian estimation of concordance among gene trees. Mol. Biol. Evol. 2007;24:412–426. - PubMed
-
- Armbruster S., Edwards M., Debevec E. Floral character displacement generates assemblage structure of Western Australian triggerplants (Stylidium) Ecology. 1994;75:315–329.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

