G-quadruplex structures are stable and detectable in human genomic DNA
- PMID: 23653208
- PMCID: PMC3736099
- DOI: 10.1038/ncomms2792
G-quadruplex structures are stable and detectable in human genomic DNA
Abstract
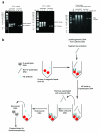
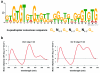
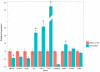
The G-quadruplex is an alternative DNA structural motif that is considered to be functionally important in the mammalian genome for transcriptional regulation, DNA replication and genome stability, but the nature and distribution of G-quadruplexes across the genome remains elusive. Here, we address the hypothesis that G-quadruplex structures exist within double-stranded genomic DNA and can be explicitly identified using a G-quadruplex-specific probe. An engineered antibody is employed to enrich for DNA containing G-quadruplex structures, followed by deep sequencing to detect and map G-quadruplexes at high resolution in genomic DNA from human breast adenocarcinoma cells. Our high sensitivity structure-based pull-down strategy enables the isolation of genomic DNA fragments bearing single, as well as multiple G-quadruplex structures. Stable G-quadruplex structures are found in sub-telomeres, gene bodies and gene regulatory regions. For a sample of identified target genes, we show that G-quadruplex-stabilizing ligands can modulate transcription. These results confirm the existence of G-quadruplex structures and their persistence in human genomic DNA.
Figures

References
-
- Verma A, et al. Genome-wide computational and expression analyses reveal G-quadruplex DNA motifs as conserved cis-regulatory elements in human and related species. J Med Chem. 2008;51:5641–5649. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

