Hereditary Hemorrhagic Telangiectasia: Breakpoint Characterization of a Novel Large Deletion in ACVRL1 Suggests the Causing Mechanism
- PMID: 23653583
- PMCID: PMC3638981
- DOI: 10.1159/000347029
Hereditary Hemorrhagic Telangiectasia: Breakpoint Characterization of a Novel Large Deletion in ACVRL1 Suggests the Causing Mechanism
Abstract
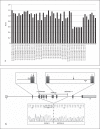
Hereditary hemorrhagic telangiectasia (HHT) is an autosomal dominant vascular dysplasia. Mutations in either ENG or ACVRL1 account for around 85% of cases, and 10% are large deletions and duplications. Here we present a large novel deletion in ACVRL1 gene and its molecular characterization in a 3 generation Italian family. We employed short tandem repeats (STRs) analysis, direct sequencing, multiplex ligation-dependant probe amplification (MLPA) analysis, and 'deletion-specific' PCR methods. STRs Analysis at ENG and ACVRL1 loci suggested a positive linkage for ACVRL1. Direct sequencing of this gene did not identify any mutations, while MLPA identified a large deletion. These results were confirmed and exactly characterized with a 'deletion-specific' PCR: the deletion size is 4,594 bp and breakpoints in exon 3 and intron 8 show the presence of short direct repeats of 7 bp [GCCCCAC]. We hypothesize, as causative molecular mechanism, the replication slippage model. Understanding the fine mechanisms associated with genomic rearrangements may indicate the nonrandomness of these events, highlighting hot spots regions. The complete concordance among MLPA, STRs analysis and 'deletion-specific PCR' supports the usefulness of MLPA in HHT molecular analysis.
Keywords: ACVRL1; Large deletion; MLPA; Short direct repeats; Slippage.
Figures

References
-
- Abeysinghe SS, Chuzhanova N, Krawczak M, Ball EV, Cooper DN. Translocation and gross deletion breakpoints in human inherited disease and cancer I: nucleotide composition and recombination-associated motifs. Hum Mutat. 2003;22:229–244. - PubMed
-
- Bacolla A, Wojciechowska M, Kosmider B, Larson JE, Wells RD. The involvement of non-B DNA structures in gross chromosomal rearrangements. DNA Repair (Amst) 2006;5:1161–1170. - PubMed
-
- Bayrak-Toydemir P, McDonald J, Akarsu N, Toydemir RM, Calderon F, et al. A fourth locus for hereditary hemorrhagic telangiectasia maps to chromosome 7. Am J Med Genet A. 2006;140:2155–2162. - PubMed
-
- Chen JM, Chuzhanova N, Stenson PD, Ferec C, Cooper DN. Complex gene rearrangements caused by serial replication slippage. Hum Mutat. 2005a;26:125–134. - PubMed
-
- Chen JM, Chuzhanova N, Stenson PD, Ferec C, Cooper DN. Meta-analysis of gross insertions causing human genetic disease: novel mutational mechanisms and the role of replication slippage. Hum Mutat. 2005b;25:207–221. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

