The YNP Metagenome Project: Environmental Parameters Responsible for Microbial Distribution in the Yellowstone Geothermal Ecosystem
- PMID: 23653623
- PMCID: PMC3644721
- DOI: 10.3389/fmicb.2013.00067
The YNP Metagenome Project: Environmental Parameters Responsible for Microbial Distribution in the Yellowstone Geothermal Ecosystem
Abstract
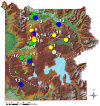
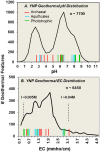
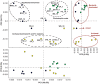
The Yellowstone geothermal complex contains over 10,000 diverse geothermal features that host numerous phylogenetically deeply rooted and poorly understood archaea, bacteria, and viruses. Microbial communities in high-temperature environments are generally less diverse than soil, marine, sediment, or lake habitats and therefore offer a tremendous opportunity for studying the structure and function of different model microbial communities using environmental metagenomics. One of the broader goals of this study was to establish linkages among microbial distribution, metabolic potential, and environmental variables. Twenty geochemically distinct geothermal ecosystems representing a broad spectrum of Yellowstone hot-spring environments were used for metagenomic and geochemical analysis and included approximately equal numbers of: (1) phototrophic mats, (2) "filamentous streamer" communities, and (3) archaeal-dominated sediments. The metagenomes were analyzed using a suite of complementary and integrative bioinformatic tools, including phylogenetic and functional analysis of both individual sequence reads and assemblies of predominant phylotypes. This volume identifies major environmental determinants of a large number of thermophilic microbial lineages, many of which have not been fully described in the literature nor previously cultivated to enable functional and genomic analyses. Moreover, protein family abundance comparisons and in-depth analyses of specific genes and metabolic pathways relevant to these hot-spring environments reveal hallmark signatures of metabolic capabilities that parallel the distribution of phylotypes across specific types of geochemical environments.
Keywords: functional genomics; geochemistry; microbial interactions; microbial mats; thermophiles.
Figures





References
-
- Amend J. P., Rogers K. L., Shock E. L., Gurrieri S., Inguaggiato S. (2003). Energetics of chemolithotrophy in the hydrothermal system of Vulcano Island, southern Italy. Geobiology 1, 37–58 10.1046/j.1472-4669.2003.00006.x - DOI
-
- Badger J. H., Hoover T. R., Brun Y. V., Weiner R. M., Laub M. T., Alexandre G., et al. (2006). Comparative genomic evidence for a close relationship between the dimorphic prosthecate bacteria Hyphomonas neptunium and Caulobacter crescentus. J. Bacteriol. 188, 6841–6850 10.1128/JB.00111-06 - DOI - PMC - PubMed
-
- Ball J. W., McCleskey R. B., Nordstrom D. K. (2010). Water-chemistry Data for Selected Springs, Geysers, and Streams in Yellowstone National Park, Wyoming, 2006-2008. U.S. Geological Survey Open-File Report 2010-1192, 109 Available at: http://pubs.usgs.gov/of/2010/1192/
LinkOut - more resources
Full Text Sources
Other Literature Sources

