Opportunities for synthetic biology in antibiotics: expanding glycopeptide chemical diversity
- PMID: 23654249
- PMCID: PMC4384835
- DOI: 10.1021/sb300092n
Opportunities for synthetic biology in antibiotics: expanding glycopeptide chemical diversity
Abstract
Synthetic biology offers a new path for the exploitation and improvement of natural products to address the growing crisis in antibiotic resistance. All antibiotics in clinical use are facing eventual obsolesce as a result of the evolution and dissemination of resistance mechanisms, yet there are few new drug leads forthcoming from the pharmaceutical sector. Natural products of microbial origin have proven over the past 70 years to be the wellspring of antimicrobial drugs. Harnessing synthetic biology thinking and strategies can provide new molecules and expand chemical diversity of known antibiotic scaffolds to provide much needed new drug leads. The glycopeptide antibiotics offer paradigmatic scaffolds suitable for such an approach. We review these strategies here using the glycopeptides as an example and demonstrate how synthetic biology can expand antibiotic chemical diversity to help address the growing resistance crisis.
Keywords: antibiotic scaffold; glycopeptides; heterologous host; resistance; synthetic biology; tailoring enzymes.
Figures

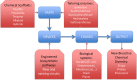
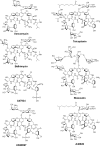


References
-
- Walsh C. T. (2003) Antibiotics: Actions, Origins, Resistance, ASM press, Washington, DC.
-
- Miller S. J.; Clardy J. (2009) Natural products: Beyond grind and find. Nat. Chem. 1, 261–263. - PubMed
-
- Li J. W.; Vederas J. C. (2009) Drug discovery and natural products: end of an era or an endless frontier?. Science 325, 161–165. - PubMed
-
- Wright G. D. (2007) The antibiotic resistome: the nexus of chemical and genetic diversity. Nat. Rev. Microbiol. 5, 175–186. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

