Molecular mechanisms of RNA interference
- PMID: 23654304
- PMCID: PMC5895182
- DOI: 10.1146/annurev-biophys-083012-130404
Molecular mechanisms of RNA interference
Abstract
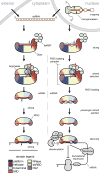
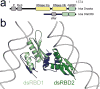
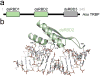
Small RNA molecules regulate eukaryotic gene expression during development and in response to stresses including viral infection. Specialized ribonucleases and RNA-binding proteins govern the production and action of small regulatory RNAs. After initial processing in the nucleus by Drosha, precursor microRNAs (pre-miRNAs) are transported to the cytoplasm, where Dicer cleavage generates mature microRNAs (miRNAs) and short interfering RNAs (siRNAs). These double-stranded products assemble with Argonaute proteins such that one strand is preferentially selected and used to guide sequence-specific silencing of complementary target mRNAs by endonucleolytic cleavage or translational repression. Molecular structures of Dicer and Argonaute proteins, and of RNA-bound complexes, have offered exciting insights into the mechanisms operating at the heart of RNA-silencing pathways.
Figures

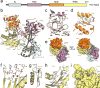
References
-
- Bellemer C, Bortolin-Cavaillé M-L, Schmidt U, Jensen SMR, Kjems J, Bertrand E, Cavaillé J. Microprocessor dynamics and interactions at endogenous imprinted C19MC microRNA genes. J. Cell. Sci. 2012;125(Pt 11):2709–2720. - PubMed
-
- Berezikov E. Nat. Rev. Genet. 12. Vol. 12. Nature Publishing Group; 2011. Evolution of microRNA diversity and regulation in animals; pp. 846–860. - PubMed
-
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409(6818):363–366. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

