Genetic circuit performance under conditions relevant for industrial bioreactors
- PMID: 23656232
- PMCID: PMC3904225
- DOI: 10.1021/sb3000832
Genetic circuit performance under conditions relevant for industrial bioreactors
Abstract
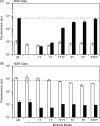
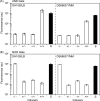
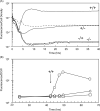
Synthetic genetic programs promise to enable novel applications in industrial processes. For such applications, the genetic circuits that compose programs will require fidelity in varying and complex environments. In this work, we report the performance of two synthetic circuits in Escherichia coli under industrially relevant conditions, including the selection of media, strain, and growth rate. We test and compare two transcriptional circuits: an AND and a NOR gate. In E. coli DH10B, the AND gate is inactive in minimal media; activity can be rescued by supplementing the media and transferring the gate into the industrial strain E. coli DS68637 where normal function is observed in minimal media. In contrast, the NOR gate is robust to media composition and functions similarly in both strains. The AND gate is evaluated at three stages of early scale-up: 100 mL shake flask experiments, a 1 mL MTP microreactor, and a 10 L bioreactor. A reference plasmid that constitutively produces a GFP reporter is used to make comparisons of circuit performance across conditions. The AND gate function is quantitatively different at each scale. The output deteriorates late in fermentation after the shift from exponential to constant feed rates, which induces rapid resource depletion and changes in growth rate. In addition, one of the output states of the AND gate failed in the bioreactor, effectively making it only responsive to a single input. Finally, cells carrying the AND gate show considerably less accumulation of biomass. Overall, these results highlight challenges and suggest modified strategies for developing and characterizing genetic circuits that function reliably during fermentation.
Figures





References
-
- Medema MH, Breitling R, Bovenberg R, Takano E. Exploiting plug-and-play synthetic biology for drug discovery and production in microorganisms. Nat Rev Microbiol. 2011;9:131–137. - PubMed
-
- Anderson JC, Clarke EJ, Arkin AP, Voigt CA. Environmentally controlled invasion of cancer cells by engineered bacteria. J Mol Biol. 2006;355:619–627. - PubMed
-
- Purnick PEM, Weiss R. The second wave of synthetic biology: from modules to systems. Nat Rev. 2009;10:410–422. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

