Acetylation dynamics of human nuclear proteins during the ionizing radiation-induced DNA damage response
- PMID: 23656789
- PMCID: PMC3713127
- DOI: 10.4161/cc.24758
Acetylation dynamics of human nuclear proteins during the ionizing radiation-induced DNA damage response
Abstract
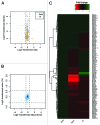
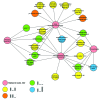
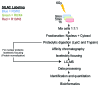
Genotoxic insults, such as ionizing radiation (IR), cause DNA damage that evokes a multifaceted cellular DNA damage response (DDR). DNA damage signaling events that control protein activity, subcellular localization, DNA binding, protein-protein interactions, etc. rely heavily on time-dependent posttranslational modifications (PTMs). To complement our previous analysis of IR-induced temporal dynamics of nuclear phosphoproteome, we now identify a range of human nuclear proteins that are dynamically regulated by acetylation, and predominantly deacetylation, during IR-induced DDR by using mass spectrometry-based proteomic approaches. Apart from cataloging acetylation sites through SILAC proteomic analyses before IR and at 5 and 60 min after IR exposure of U2OS cells, we report that: (1) key components of the transcriptional machinery, such as EP300 and CREBBP, are dynamically acetylated; (2) that nuclear acetyltransferases themselves are regulated, not on the protein abundance level, but by (de)acetylation; and (3) that the recently reported p53 co-activator and methyltransferase MLL3 is acetylated on five lysines during the DDR. For selected examples, protein immunoprecipitation and immunoblotting were used to assess lysine acetylation status and thereby validate the mass spectrometry data. We thus present evidence that nuclear proteins, including those known to regulate cellular functions via epigenetic modifications of histones, are regulated by (de)acetylation in a timely manner upon cell's exposure to genotoxic insults. Overall, these results present a resource of temporal profiles of a spectrum of protein acetylation sites during DDR and provide further insights into the highly dynamic nature of regulatory PTMs that help orchestrate the maintenance of genome integrity.
Keywords: DNA damage response; ionizing radiation; nucleus; protein acetylation; quantitative proteomics.
Figures

Comment in
-
Rapid and transient protein acetylation changes in response to DNA damage.Cell Cycle. 2013 Jul 1;12(13):1993. doi: 10.4161/cc.25315. Epub 2013 Jun 11. Cell Cycle. 2013. PMID: 23759576 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
