Exploring the three-dimensional organization of genomes: interpreting chromatin interaction data
- PMID: 23657480
- PMCID: PMC3874835
- DOI: 10.1038/nrg3454
Exploring the three-dimensional organization of genomes: interpreting chromatin interaction data
Abstract
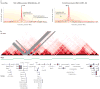
How DNA is organized in three dimensions inside the cell nucleus and how this affects the ways in which cells access, read and interpret genetic information are among the longest standing questions in cell biology. Using newly developed molecular, genomic and computational approaches based on the chromosome conformation capture technology (such as 3C, 4C, 5C and Hi-C), the spatial organization of genomes is being explored at unprecedented resolution. Interpreting the increasingly large chromatin interaction data sets is now posing novel challenges. Here we describe several types of statistical and computational approaches that have recently been developed to analyse chromatin interaction data.
Figures
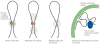



References
-
- Dekker J, Rippe K, Dekker M, Kleckner N. Capturing Chromosome Conformation. Science. 2002;295:1306–1311. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

