Affinity purification of T7 RNA transcripts with homogeneous ends using ARiBo and CRISPR tags
- PMID: 23657939
- PMCID: PMC3683919
- DOI: 10.1261/rna.037432.112
Affinity purification of T7 RNA transcripts with homogeneous ends using ARiBo and CRISPR tags
Abstract
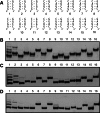
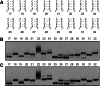
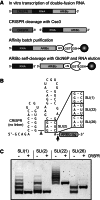
Affinity purification of RNA using the ARiBo tag technology currently provides an ideal approach to quickly prepare RNA with 3' homogeneity. Here, we explored strategies to also ensure 5' homogeneity of affinity-purified RNAs. First, we systematically investigated the effect of starting nucleotides on the 5' heterogeneity of a small SLI RNA substrate from the Neurospora VS ribozyme purified from an SLI-ARiBo precursor. A series of 32 SLI RNA sequences with variations in the +1 to +3 region was produced from two T7 promoters (class III consensus and class II 2.5) using either the wild-type T7 RNA polymerase or the P266L mutant. Although the P266L mutant helps decrease the levels of 5'-sequence heterogeneity in several cases, significant levels of 5' heterogeneity (≥1.5%) remain for transcripts starting with GGG, GAG, GCG, GGC, AGG, AGA, AAA, ACA, AUA, AAC, ACC, AUC, and AAU. To provide a more general approach to purifying RNA with 5' homogeneity, we tested the suitability of using a small CRISPR RNA stem-loop at the 5' end of the SLI-ARiBo RNA. Interestingly, we found that complete cleavage of the 5'-CRISPR tag with the Cse3 endoribonuclease can be achieved quickly from CRISPR-SLI-ARiBo transcripts. With this procedure, it is possible to generate SLI-ARiBo RNAs starting with any of the four standard nucleotides (G, C, A, or U) involved in either a single- or a double-stranded structure. Moreover, the 5'-CRISPR-based strategy can be combined with affinity purification using the 3'-ARiBo tag for quick purification of RNA with both 5' and 3' homogeneity.
Keywords: 5′ heterogeneity; ARiBo tag; CRISPR tag; Cse3 endoribonuclease; T7 RNA polymerase; affinity purification of RNA.
Figures


References
-
- Al-Attar S, Westra ER, van der Oost J, Brouns SJ 2011. Clustered regularly interspaced short palindromic repeats (CRISPRs): The hallmark of an ingenious antiviral defense mechanism in prokaryotes. Biol Chem 392: 277–289 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
