The impact of trans-regulation on the evolutionary rates of metazoan proteins
- PMID: 23658220
- PMCID: PMC3711421
- DOI: 10.1093/nar/gkt349
The impact of trans-regulation on the evolutionary rates of metazoan proteins
Abstract
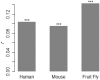
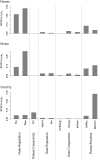
Transcription factor (TF) and microRNA (miRNA) are two crucial trans-regulatory factors that coordinately control gene expression. Understanding the impacts of these two factors on the rate of protein sequence evolution is of great importance in evolutionary biology. While many biological factors associated with evolutionary rate variations have been studied, evolutionary analysis of simultaneously accounting for TF and miRNA regulations across metazoans is still uninvestigated. Here, we provide a series of statistical analyses to assess the influences of TF and miRNA regulations on evolutionary rates across metazoans (human, mouse and fruit fly). Our results reveal that the negative correlations between trans-regulation and evolutionary rates hold well across metazoans, but the strength of TF regulation as a rate indicator becomes weak when the other confounding factors that may affect evolutionary rates are controlled. We show that miRNA regulation tends to be a more essential indicator of evolutionary rates than TF regulation, and the combination of TF and miRNA regulations has a significant dependent effect on protein evolutionary rates. We also show that trans-regulation (especially miRNA regulation) is much more important in human/mouse than in fruit fly in determining protein evolutionary rates, suggesting a considerable variation in rate determinants between vertebrates and invertebrates.
Figures
Similar articles
-
Impacts of pretranscriptional DNA methylation, transcriptional transcription factor, and posttranscriptional microRNA regulations on protein evolutionary rate.Genome Biol Evol. 2014 Jun 12;6(6):1530-41. doi: 10.1093/gbe/evu124. Genome Biol Evol. 2014. PMID: 24923326 Free PMC article.
-
MicroRNA networks alter to conform to transcription factor networks adding redundancy and reducing the repertoire of target genes for coordinated regulation.Mol Biol Evol. 2011 Jan;28(1):639-46. doi: 10.1093/molbev/msq231. Epub 2010 Aug 30. Mol Biol Evol. 2011. PMID: 20805189
-
Coordinated networks of microRNAs and transcription factors with evolutionary perspectives.Adv Exp Med Biol. 2013;774:169-87. doi: 10.1007/978-94-007-5590-1_10. Adv Exp Med Biol. 2013. PMID: 23377974 Review.
-
DIANA-mirExTra v2.0: Uncovering microRNAs and transcription factors with crucial roles in NGS expression data.Nucleic Acids Res. 2016 Jul 8;44(W1):W128-34. doi: 10.1093/nar/gkw455. Epub 2016 May 20. Nucleic Acids Res. 2016. PMID: 27207881 Free PMC article.
-
Evolutionary conservation of microRNA regulatory circuits: an examination of microRNA gene complexity and conserved microRNA-target interactions through metazoan phylogeny.DNA Cell Biol. 2007 Apr;26(4):209-18. doi: 10.1089/dna.2006.0545. DNA Cell Biol. 2007. PMID: 17465887 Review.
Cited by
-
The evolution of the coding exome of the Arabidopsis species--the influences of DNA methylation, relative exon position, and exon length.BMC Evol Biol. 2014 Jun 25;14:145. doi: 10.1186/1471-2148-14-145. BMC Evol Biol. 2014. PMID: 24965500 Free PMC article.
-
Impacts of pretranscriptional DNA methylation, transcriptional transcription factor, and posttranscriptional microRNA regulations on protein evolutionary rate.Genome Biol Evol. 2014 Jun 12;6(6):1530-41. doi: 10.1093/gbe/evu124. Genome Biol Evol. 2014. PMID: 24923326 Free PMC article.
-
Purifying selection shapes the coincident SNP distribution of primate coding sequences.Sci Rep. 2016 Jun 3;6:27272. doi: 10.1038/srep27272. Sci Rep. 2016. PMID: 27255481 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

