A poor man's BLASTX--high-throughput metagenomic protein database search using PAUDA
- PMID: 23658416
- PMCID: PMC3866550
- DOI: 10.1093/bioinformatics/btt254
A poor man's BLASTX--high-throughput metagenomic protein database search using PAUDA
Abstract
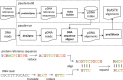
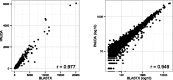
Summary: In the context of metagenomics, we introduce a new approach to protein database search called PAUDA, which runs ~10,000 times faster than BLASTX, while achieving about one-third of the assignment rate of reads to KEGG orthology groups, and producing gene and taxon abundance profiles that are highly correlated to those obtained with BLASTX. PAUDA requires <80 CPU hours to analyze a dataset of 246 million Illumina DNA reads from permafrost soil for which a previous BLASTX analysis (on a subset of 176 million reads) reportedly required 800,000 CPU hours, leading to the same clustering of samples by functional profiles.
Availability: PAUDA is freely available from: http://ab.inf.uni-tuebingen.de/software/pauda. Also supplementary method details are available from this website.
Figures
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

