Phosphorylation of CDK9 at Ser175 enhances HIV transcription and is a marker of activated P-TEFb in CD4(+) T lymphocytes
- PMID: 23658523
- PMCID: PMC3642088
- DOI: 10.1371/journal.ppat.1003338
Phosphorylation of CDK9 at Ser175 enhances HIV transcription and is a marker of activated P-TEFb in CD4(+) T lymphocytes
Abstract
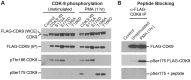
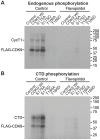
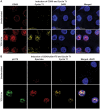
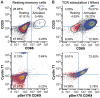
The HIV transactivator protein, Tat, enhances HIV transcription by recruiting P-TEFb from the inactive 7SK snRNP complex and directing it to proviral elongation complexes. To test the hypothesis that T-cell receptor (TCR) signaling induces critical post-translational modifications leading to enhanced interactions between P-TEFb and Tat, we employed affinity purification-tandem mass spectrometry to analyze P-TEFb. TCR or phorbal ester (PMA) signaling strongly induced phosphorylation of the CDK9 kinase at Ser175. Molecular modeling studies based on the Tat/P-TEFb X-ray structure suggested that pSer175 strengthens the intermolecular interactions between CDK9 and Tat. Mutations in Ser175 confirm that this residue could mediate critical interactions with Tat and with the bromodomain protein BRD4. The S175A mutation reduced CDK9 interactions with Tat by an average of 1.7-fold, but also completely blocked CDK9 association with BRD4. The phosphomimetic S175D mutation modestly enhanced Tat association with CDK9 while causing a 2-fold disruption in BRD4 association with CDK9. Since BRD4 is unable to compete for binding to CDK9 carrying S175A, expression of CDK9 carrying the S175A mutation in latently infected cells resulted in a robust Tat-dependent reactivation of the provirus. Similarly, the stable knockdown of BRD4 led to a strong enhancement of proviral expression. Immunoprecipitation experiments show that CDK9 phosphorylated at Ser175 is excluded from the 7SK RNP complex. Immunofluorescence and flow cytometry studies carried out using a phospho-Ser175-specific antibody demonstrated that Ser175 phosphorylation occurs during TCR activation of primary resting memory CD4+ T cells together with upregulation of the Cyclin T1 regulatory subunit of P-TEFb, and Thr186 phosphorylation of CDK9. We conclude that the phosphorylation of CDK9 at Ser175 plays a critical role in altering the competitive binding of Tat and BRD4 to P-TEFb and provides an informative molecular marker for the identification of the transcriptionally active form of P-TEFb.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures








Similar articles
-
Cyclin-dependent kinase 7 (CDK7)-mediated phosphorylation of the CDK9 activation loop promotes P-TEFb assembly with Tat and proviral HIV reactivation.J Biol Chem. 2018 Jun 29;293(26):10009-10025. doi: 10.1074/jbc.RA117.001347. Epub 2018 May 9. J Biol Chem. 2018. PMID: 29743242 Free PMC article.
-
T-loop phosphorylated Cdk9 localizes to nuclear speckle domains which may serve as sites of active P-TEFb function and exchange between the Brd4 and 7SK/HEXIM1 regulatory complexes.J Cell Physiol. 2010 Jul;224(1):84-93. doi: 10.1002/jcp.22096. J Cell Physiol. 2010. PMID: 20201073 Free PMC article.
-
T-cell receptor signaling enhances transcriptional elongation from latent HIV proviruses by activating P-TEFb through an ERK-dependent pathway.J Mol Biol. 2011 Jul 29;410(5):896-916. doi: 10.1016/j.jmb.2011.03.054. J Mol Biol. 2011. PMID: 21763495 Free PMC article.
-
The HIV-1 Tat Protein: Mechanism of Action and Target for HIV-1 Cure Strategies.Curr Pharm Des. 2017;23(28):4098-4102. doi: 10.2174/1381612823666170704130635. Curr Pharm Des. 2017. PMID: 28677507 Free PMC article. Review.
-
Regulation of TAK/P-TEFb in CD4+ T lymphocytes and macrophages.Curr HIV Res. 2003 Oct;1(4):395-404. doi: 10.2174/1570162033485159. Curr HIV Res. 2003. PMID: 15049426 Review.
Cited by
-
Viral-host interactions that control HIV-1 transcriptional elongation.Chem Rev. 2013 Nov 13;113(11):8567-82. doi: 10.1021/cr400120z. Epub 2013 Jun 24. Chem Rev. 2013. PMID: 23795863 Free PMC article. Review.
-
Evolutionary distance of amino acid sequence orthologs across macaque subspecies: identifying candidate genes for SIV resistance in Chinese rhesus macaques.PLoS One. 2015 Apr 17;10(4):e0123624. doi: 10.1371/journal.pone.0123624. eCollection 2015. PLoS One. 2015. PMID: 25884674 Free PMC article.
-
MD simulation of the Tat/Cyclin T1/CDK9 complex revealing the hidden catalytic cavity within the CDK9 molecule upon Tat binding.PLoS One. 2017 Feb 8;12(2):e0171727. doi: 10.1371/journal.pone.0171727. eCollection 2017. PLoS One. 2017. PMID: 28178316 Free PMC article.
-
Efficient Non-Epigenetic Activation of HIV Latency through the T-Cell Receptor Signalosome.Viruses. 2020 Aug 8;12(8):868. doi: 10.3390/v12080868. Viruses. 2020. PMID: 32784426 Free PMC article. Review.
-
Transcriptional control of HIV latency: cellular signaling pathways, epigenetics, happenstance and the hope for a cure.Virology. 2014 Apr;454-455:328-39. doi: 10.1016/j.virol.2014.02.008. Epub 2014 Feb 22. Virology. 2014. PMID: 24565118 Free PMC article. Review.
References
-
- Richman DD, Margolis DM, Delaney M, Greene WC, Hazuda D, et al. (2009) The challenge of finding a cure for HIV infection. Science 323: 1304–1307. - PubMed
-
- Han Y, Wind-Rotolo M, Yang HC, Siliciano JD, Siliciano RF (2007) Experimental approaches to the study of HIV-1 latency. Nat Rev Microbiol 5: 95–106. - PubMed
-
- Margolis DM (2010) Mechanisms of HIV latency: an emerging picture of complexity. Curr HIV/AIDS Rep 7: 37–43. - PubMed
-
- Trono D, Van Lint C, Rouzioux C, Verdin E, Barre-Sinoussi F, et al. (2010) HIV persistence and the prospect of long-term drug-free remissions for HIV-infected individuals. Science 329: 174–180. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

