Chromosomal organization and segregation in Pseudomonas aeruginosa
- PMID: 23658532
- PMCID: PMC3642087
- DOI: 10.1371/journal.pgen.1003492
Chromosomal organization and segregation in Pseudomonas aeruginosa
Abstract
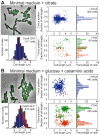
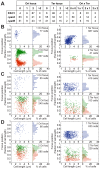
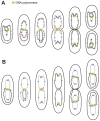
The study of chromosomal organization and segregation in a handful of bacteria has revealed surprising variety in the mechanisms mediating such fundamental processes. In this study, we further emphasized this diversity by revealing an original organization of the Pseudomonas aeruginosa chromosome. We analyzed the localization of 20 chromosomal markers and several components of the replication machinery in this important opportunistic γ-proteobacteria pathogen. This technique allowed us to show that the 6.3 Mb unique circular chromosome of P. aeruginosa is globally oriented from the old pole of the cell to the division plane/new pole along the oriC-dif axis. The replication machinery is positioned at mid-cell, and the chromosomal loci from oriC to dif are moved sequentially to mid-cell prior to replication. The two chromosomal copies are subsequently segregated at their final subcellular destination in the two halves of the cell. We identified two regions in which markers localize at similar positions, suggesting a bias in the distribution of chromosomal regions in the cell. The first region encompasses 1.4 Mb surrounding oriC, where loci are positioned around the 0.2/0.8 relative cell length upon segregation. The second region contains at least 800 kb surrounding dif, where loci show an extensive colocalization step following replication. We also showed that disrupting the ParABS system is very detrimental in P. aeruginosa. Possible mechanisms responsible for the coordinated chromosomal segregation process and for the presence of large distinctive regions are discussed.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Regional Control of Chromosome Segregation in Pseudomonas aeruginosa.PLoS Genet. 2016 Nov 7;12(11):e1006428. doi: 10.1371/journal.pgen.1006428. eCollection 2016 Nov. PLoS Genet. 2016. PMID: 27820816 Free PMC article.
-
Segregation but Not Replication of the Pseudomonas aeruginosa Chromosome Terminates at Dif.mBio. 2018 Oct 23;9(5):e01088-18. doi: 10.1128/mBio.01088-18. mBio. 2018. PMID: 30352930 Free PMC article.
-
Alternating Dynamics of oriC, SMC, and MksBEF in Segregation of Pseudomonas aeruginosa Chromosome.mSphere. 2020 Sep 9;5(5):e00238-20. doi: 10.1128/mSphere.00238-20. mSphere. 2020. PMID: 32907947 Free PMC article.
-
Maintenance of chromosome structure in Pseudomonas aeruginosa.FEMS Microbiol Lett. 2014 Jul;356(2):154-65. doi: 10.1111/1574-6968.12478. Epub 2014 Jun 12. FEMS Microbiol Lett. 2014. PMID: 24863732 Free PMC article. Review.
-
Escherichia coli and its chromosome.Trends Microbiol. 2008 May;16(5):238-45. doi: 10.1016/j.tim.2008.02.003. Epub 2008 Apr 9. Trends Microbiol. 2008. PMID: 18406139 Review.
Cited by
-
Differential management of the replication terminus regions of the two Vibrio cholerae chromosomes during cell division.PLoS Genet. 2014 Sep 25;10(9):e1004557. doi: 10.1371/journal.pgen.1004557. eCollection 2014 Sep. PLoS Genet. 2014. PMID: 25255436 Free PMC article.
-
Post-replicative pairing of sister ter regions in Escherichia coli involves multiple activities of MatP.Nat Commun. 2020 Jul 30;11(1):3796. doi: 10.1038/s41467-020-17606-6. Nat Commun. 2020. PMID: 32732900 Free PMC article.
-
Regional Control of Chromosome Segregation in Pseudomonas aeruginosa.PLoS Genet. 2016 Nov 7;12(11):e1006428. doi: 10.1371/journal.pgen.1006428. eCollection 2016 Nov. PLoS Genet. 2016. PMID: 27820816 Free PMC article.
-
Stable Regulation of Cell Cycle Events in Mycobacteria: Insights From Inherently Heterogeneous Bacterial Populations.Front Microbiol. 2018 Mar 21;9:514. doi: 10.3389/fmicb.2018.00514. eCollection 2018. Front Microbiol. 2018. PMID: 29619019 Free PMC article. Review.
-
New approaches to understanding the spatial organization of bacterial genomes.Curr Opin Microbiol. 2014 Dec;22:15-21. doi: 10.1016/j.mib.2014.09.014. Epub 2014 Oct 7. Curr Opin Microbiol. 2014. PMID: 25305533 Free PMC article. Review.
References
-
- Silby MW, Winstanley C, Godfrey SA, Levy SB, Jackson RW (2011) Pseudomonas genomes: diverse and adaptable. FEMS Microbiol Rev 35: 652–680. - PubMed
-
- Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, et al. (2000) Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406: 959–964. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

