Within-host evolution of Staphylococcus aureus during asymptomatic carriage
- PMID: 23658690
- PMCID: PMC3641031
- DOI: 10.1371/journal.pone.0061319
Within-host evolution of Staphylococcus aureus during asymptomatic carriage
Abstract
Background: Staphylococcus aureus is a major cause of healthcare associated mortality, but like many important bacterial pathogens, it is a common constituent of the normal human body flora. Around a third of healthy adults are carriers. Recent evidence suggests that evolution of S. aureus during nasal carriage may be associated with progression to invasive disease. However, a more detailed understanding of within-host evolution under natural conditions is required to appreciate the evolutionary and mechanistic reasons why commensal bacteria such as S. aureus cause disease. Therefore we examined in detail the evolutionary dynamics of normal, asymptomatic carriage. Sequencing a total of 131 genomes across 13 singly colonized hosts using the Illumina platform, we investigated diversity, selection, population dynamics and transmission during the short-term evolution of S. aureus.
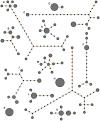
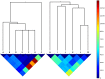
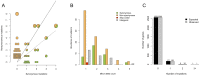
Principal findings: We characterized the processes by which the raw material for evolution is generated: micro-mutation (point mutation and small insertions/deletions), macro-mutation (large insertions/deletions) and the loss or acquisition of mobile elements (plasmids and bacteriophages). Through an analysis of synonymous, non-synonymous and intergenic mutations we discovered a fitness landscape dominated by purifying selection, with rare examples of adaptive change in genes encoding surface-anchored proteins and an enterotoxin. We found evidence for dramatic, hundred-fold fluctuations in the size of the within-host population over time, which we related to the cycle of colonization and clearance. Using a newly-developed population genetics approach to detect recent transmission among hosts, we revealed evidence for recent transmission between some of our subjects, including a husband and wife both carrying populations of methicillin-resistant S. aureus (MRSA).
Significance: This investigation begins to paint a picture of the within-host evolution of an important bacterial pathogen during its prevailing natural state, asymptomatic carriage. These results also have wider significance as a benchmark for future systematic studies of evolution during invasive S. aureus disease.
Conflict of interest statement
Figures



References
-
- Wertheim HF, Melles DC, Vos MC, van Leeuwen W, van Belkum A, et al. (2005) The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect Dis 5: 751–762. - PubMed
-
- World Health Organization (2008) The Global Burden of Disease: 2004 Update. Available: http://www.who.int/healthinfo/global_burden_disease. Accessed 2012 April 12.
-
- World Health Organization (2011) World Health Organization Mortality Database, ICD-10. 24 November 2011 update. Available: http://www.who.int/whosis/mort/download/en/index.html. Accessed 2012 April 12.
-
- Moxon ER, Jansen VAA (2005) Phage variation: understanding the behaviour of an accidental pathogen. Trends Microbiol 13: 563–565. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

