Comparative transcriptional profiling of the axolotl limb identifies a tripartite regeneration-specific gene program
- PMID: 23658691
- PMCID: PMC3641036
- DOI: 10.1371/journal.pone.0061352
Comparative transcriptional profiling of the axolotl limb identifies a tripartite regeneration-specific gene program
Abstract
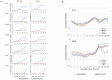
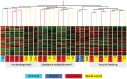
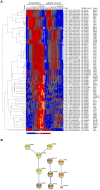
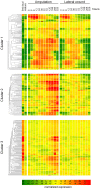
Understanding how the limb blastema is established after the initial wound healing response is an important aspect of regeneration research. Here we performed parallel expression profile time courses of healing lateral wounds versus amputated limbs in axolotl. This comparison between wound healing and regeneration allowed us to identify amputation-specific genes. By clustering the expression profiles of these samples, we could detect three distinguishable phases of gene expression - early wound healing followed by a transition-phase leading to establishment of the limb development program, which correspond to the three phases of limb regeneration that had been defined by morphological criteria. By focusing on the transition-phase, we identified 93 strictly amputation-associated genes many of which are implicated in oxidative-stress response, chromatin modification, epithelial development or limb development. We further classified the genes based on whether they were or were not significantly expressed in the developing limb bud. The specific localization of 53 selected candidates within the blastema was investigated by in situ hybridization. In summary, we identified a set of genes that are expressed specifically during regeneration and are therefore, likely candidates for the regulation of blastema formation.
Conflict of interest statement
Figures





References
-
- Stocum DL, Cameron JA (2011) Looking proximally and distally: 100 years of limb regeneration and beyond. Dev Dyn 240: 943–968. - PubMed
-
- Carlson BM (2003) Muscle regeneration in amphibians and mammals: passing the torch. Dev Dyn 226: 167–181. - PubMed
-
- Weiss P (1925) Unabhängigkeit der Extremitäten regeneration vom Skelett (bei Triton cristatus). Wilhelm Roux' Arch Entwicklungsmech Org 104: 359–394.
-
- Hutchison C, Pilote M, Roy S (2007) The axolotl limb: a model for bone development, regeneration and fracture healing. Bone 40: 45–56. - PubMed
-
- Satoh A, Cummings GM, Bryant SV, Gardiner DM (2010) Neurotrophic regulation of fibroblast dedifferentiation during limb skeletal regeneration in the axolotl (Ambystoma mexicanum). Dev Biol 337: 444–457. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

