Progressive impairment of muscle regeneration in muscleblind-like 3 isoform knockout mice
- PMID: 23660517
- PMCID: PMC3736872
- DOI: 10.1093/hmg/ddt209
Progressive impairment of muscle regeneration in muscleblind-like 3 isoform knockout mice
Abstract
The muscleblind-like (MBNL) genes encode alternative splicing factors that are essential for the postnatal development of multiple tissues, and the inhibition of MBNL activity by toxic C(C)UG repeat RNAs is a major pathogenic feature of the neuromuscular disease myotonic dystrophy. While MBNL1 controls fetal-to-adult splicing transitions in muscle and MBNL2 serves a similar role in the brain, the function of MBNL3 in vivo is unknown. Here, we report that mouse Mbnl3, which encodes protein isoforms that differ in the number of tandem zinc-finger RNA-binding motifs and subcellular localization, is expressed primarily during embryonic development but also transiently during injury-induced adult skeletal muscle regeneration. Mbnl3 expression is required for normal C2C12 myogenic differentiation and high-throughput sequencing combined with cross-linking/immunoprecipitation analysis indicates that Mbnl3 binds preferentially to the 3' untranslated regions of genes implicated in cell growth and proliferation. In addition, Mbnl3ΔE2 isoform knockout mice, which fail to express the major Mbnl3 nuclear isoform, show age-dependent delays in injury-induced muscle regeneration and impaired muscle function. These results suggest that Mbnl3 inhibition by toxic RNA expression may be a contributing factor to the progressive skeletal muscle weakness and wasting characteristic of myotonic dystrophy.
Figures

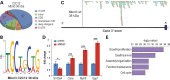
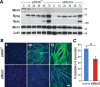

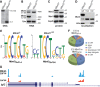
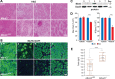
References
-
- Harper P.S. Myotonic Dystrophy. London: W.B. Saunders; 2001.
-
- Ranum L.P., Cooper T.A. RNA-mediated neuromuscular disorders. Ann. Rev. Neurosc. 2006;29:259–277. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

